Abstract
Uncontrolled retinal pigment epithelial (RPE) cell proliferation/migration contribute to the pathological tractional membrane development in proliferative vitreoretinopathy. Recent studies reported that microRNA (miR)-124 controls various cellular functions via the direct targeting of small Ras homolog family member G (RHOG). Therefore, we investigated the role of the neuron-specific miR-124 and RHOG in RPE cell proliferation/migration. Alterations in miR-124 and RhoG expression, as per cell confluence were evaluated through quantitative real-time PCR and western blotting, respectively. After transfection with miR-124, we quantified RPE cell viability and migration and observed cell polarization and lamellipodia protrusions. We evaluated the expression of RHOG/RAC1 pathway molecules in miR-124-transfected RPE cells. Endogenous miR-124 expression increased proportionally to RPE cell density, but decreased after 100% confluence. Overexpression of miR-124 decreased cell viability and migration, BrdU incorporation, and Ki-67 expression. Inhibition of endogenous miR-124 expression promoted RPE cell migration. Transfection with miR-124 reduced cell polarization, lamellipodia protrusion, and RHOG mRNA 3′ untranslated region luciferase activity. Like miR-124 overexpression, RhoG knockdown decreased RPE cell viability, wound healing, and migration, and altered the expression of cell cycle regulators. These results suggest that miR-124 could be a therapeutic target to alleviate fibrovascular proliferation in retinal diseases by regulating RPE proliferation/migration via RHOG.
Similar content being viewed by others
Introduction
Members of the small GTPase Ras homolog (RHO) family serve as molecular switches for complex cellular processes, such as migration, division, and cell cycle progression, although they are primarily known as actin cytoskeleton regulators1. Phylogenetic analysis revealed that the RHO family comprises two exclusive groups; the first group includes RHOA, B, and C, and the second includes RHOG and RAC1/22. Vincent et al. first described Ras homolog growth-related (RHOG) based on its growth-related functions2; however, the underlying functions and regulatory mechanisms of RhoG protein are largely unknown. Recently, a subset of small non-coding RNAs, microRNAs (miRNAs), were identified as endogenous regulatory RNA molecules; they play pivotal roles in the regulation of ~ 30% of all mammalian genes, mainly via repression3,4.
miR-124 is abundant in the brain and accounts for 25–48% of all central nervous system (CNS)-expressed miRNAs5,6. From the developmental perspective, the majority of CNS-specific cells begin in the neural ectoderm, including retinal pigment epithelial (RPE) cells. We hypothesized that miR-124 would be expressed in the neural retina, as it was previously identified in the murine eye by miRNA profiling7,8. The retinal pigment epithelium physiology presents unique challenges that necessitate precisely tuned regulatory networks to maintain cell viability. In steady-state conditions, RPE cells remain in the quiescent stage of the cell cycle9,10. Under pathological conditions, RPE cells are vulnerable to insults caused by the aging process and environmental stressors; proliferative vitreoretinopathy11,12, diabetic retinopathy13, and idiopathic epiretinal membranes14 can be induced by uncontrolled proliferation and RPE cell migration into vitreous space. Emerging evidence suggests that fine-tuning by miRNAs is essential for responses to cellular stress15; miRNAs play a critical role in the survival and physiology of photoreceptor cells and the retinal pigment epithelium, making them putative therapeutic targets in these blinding diseases. Furthermore, Franke et al.16 confirmed that miR-124 regulates neuronal differentiation by targeting RHOG.
In the present study, we investigated the regulatory functions of miR-124 in RPE cells; we focused particularly at cell proliferation and migration. Additionally, we assessed the physiological alterations caused by RHOG gene silencing in RPE cells, using small interfering RNAs (siRNAs).
Results
Endogenous expression levels of miR-124 and RhoG inversely correlated with RPE cell confluence
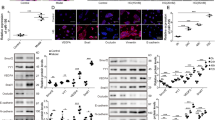
RPE cells (7.5 × 104 cells/mL) reached confluences of approximately 25%, 50%, 90%, and 100% after 24, 48, 72, and 96 h, respectively (Fig. 1A, B). The endogenous expression of miR-124 rapidly increased to threefold at 48 h compared to the level at 24 h and was maintained until 72 h after plating. However, its expression abruptly decreased at 96 h, to less than half the initial expression level, when RPE cells reached full confluence (Fig. 1C). In addition, RhoG protein expression decreased up to 72 h after plating; of note, it significantly decreased below the levels at 24, 48, and 72 h when the cells reached 100% confluence (Fig. 1D). To inhibit endogenously expressed miR-124, a miR-124-specific inhibitor was used, 24 h after RPE cell plating. As expected, the intracellular miR-124 expression levels continuously decreased up to 72 h (Fig. 1E). On the other hand, RhoG expression was increased compared to that in non-treated cells from 48 to 72 h, as per the western blot analysis (Fig. 1F).
Analysis of miR-124 and RhoG expression during cell proliferation. (A) Representative images of cell confluence after plating 7.5 × 104 RPE cells/mL in 6-well cell culture plates. (B) At the indicated time points, RPE cells were counted in triplicate cell culture wells. (C) Quantitative measurements of endogenous miR-124 expression levels in RPE cells at the indicated time points. The quantitative miRNA expression results were normalized to RNU6 and miR-16 expression levels. (D) The protein levels of cell cycle regulatory factors and RhoG were analyzed by western blotting. (E) Quantitative evaluation of endogenous miR-124 expression in non-treated or miR-124 inhibitor-treated RPE cells. (F) Expression of RhoG in non-treated or miR-124 inhibitor-treated RPE cells was assessed using western blotting.
Overexpression of miR-124 reduced RPE cell numbers and proliferative ability
After we transfected RPE cells with miR-124, we evaluated cell density through phase-contrast microscopy. The introduction of miR-124 decreased cell density, as compared with miR-NC transfection, at 24 h (Fig. 2A). We also found that miR-124 overexpression decreased cell proliferation and viability (Fig. 2B). In addition, using an absolute cell counting method, miR-124 transfection decreased cell counts by approximately 3 folds (Fig. 2C). Overexpression of miR-124 significantly decreased the number of Ki-67-positive cells within the DAPI-positive cells, as compared with mock control or miR-NC introduction (Fig. 2D, E).
Effects of miR-124 transfection on RPE cell proliferation. (A) Representative images of RPE cells after transfection with miR-124. (B) Quantitative measurement of proliferation via BrdU incorporation and viability via WST-8 assays. (C) Absolute cell counts after transfection with miR-124. (D) Ki-67 immunostaining after transfection with miR-124. (E) Percentage of Ki-67-positive cells of all cells with DAPI-positive nuclear staining. Data represent mean ± standard deviation (SD). Data analyzed by one way-ANOVA followed by Turkey’s HSD test. *p < 0.05; ***p < 0.001.
Overexpression of miR-124 impeded the motility of RPE cells
We performed in vitro wound-healing and transwell migration assays to examine the effects of miR-124 overexpression on cell motility. Overexpression of miR-124 significantly decreased the wound healing capacity of RPE cells, compared with that of miR-NC treated cells, at 24 and 48 h (Fig. 3A, B). In addition, miR-124 overexpression decreased the number of migratory cells after 12 h (Fig. 3C, D). We investigated the cause of these phenomena by evaluating the cell structure using phalloidin staining. At the leading ends of cell boundaries, miR-124-overexpressing cells lacked lamellipodia, which are crucial for cell motility. In addition, miR-124-transfected cells had altered actin structures, characterised by actin condensation at the cell border (Fig. 3E). Conversely, treatment of RPE cells with the miR-124 inhibitor, designed to antagonizse endogenous miR-124 expression, in RPE cells significantly enhanced wound healing after 16, 24, and 48 h incubations (Fig. 4A, B). In addition, miR-124 inhibitor significantly increased RPE cell migration in the transwell assay, compared with the miR inhibitor-NC (Fig. 4C, D).
Effects of miR-124 on RPE cell motility. (A) Representative images of the cross-hair wound-healing assay. (B) Quantitative measurement of the cross-hair wound after cell transfection with miR-NC or miR-124. (C) Transwell migration assay for migrating RPE cells 24 h after transfection with miR-124. (D) Statistical analysis of transfected versus control migratory RPE cells. (E) Filamentous actin staining for lamellipodium formation and cytoplasmic actin condensation. Data represent mean ± SD. Data were analyzed by independent t test. *p < 0.05; **p < 0.01.
Effects of transfection with an miR-124 inhibitor on RPE cell motility. (A) Representative images of the wound closure in monolayers of RPE cells transfected with miR inhibitor-NC and miR-124 inhibitor constructs. (B) Statistical analysis of the assay in panel (A). (C) Representative images from the transwell migration assay with RPE cells transfected with miR inhibitor-NC and miR-124 inhibitor constructs. (D) Statistical analysis of the assay in panel (C). Data represent mean ± SD. Data were analyzed by independent t test. *p < 0.05; **p < 0.01.
Identification of putative miR-124 target sequences in the RHOG 3′ UTR
To verify the regulatory effects of miR-124, we investigated potential regulatory targets with an in silico analysis using TargetScan ver. 6.2 (https://www.targetscan.org)18,19. RHOG (NM_001665) is a well-known regulatory gene related to lamellipodium formation and regulation. A search of the 3′ UTR of human RHOG revealed 2 highly evolutionarily conserved seed sequences that are targetable by miR-124 (Supplementary figure S2 A).
miR-124 directly targeted the RHOG 3′ UTR in RPE Cells
Based on an in silico analysis, in which RHOG 3′ UTR was predicted to be a target of miR-124 in RPE cells, we next performed an RHOG 3′ UTR reporter plasmid assay in RPE cells. Co-transfection of the random control reporter plasmid with miR-NC or miR-124 did not affect luciferase activity. On the other hand, co-transfection of the RHOG 3′ UTR reporter plasmid with miR-124 reduced the luciferase activity to less than 15% of that observed in control cells co-transfected with miR-NC (Supplementary figure S2 B).
Knockdown of RHOG negatively regulates cell viability and proliferation
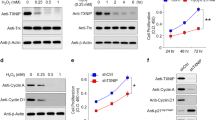
We evaluated the effects of direct RHOG silencing on RPE cell viability and proliferation. Twenty-four hours after transfection of RPE cells with si-RHOG, we observed significantly lower cell density in the transfected cells than in the control cultures (Fig. 5A). RHOG-knockdown also reduced cell viability and proliferation (Fig. 5B). In addition, we observed significantly fewer Ki-67-positive cells among the si-RHOG-transfected RPE cells than among the controls (Fig. 5C, D).
The effects of RhoG knockdown on RPE cell proliferation. (A) Representative images of RPE cells transfected with si-RhoG. (B) Quantitative analysis of the effects of RhoG knockdown on RPE cell proliferation and viability. (C) Immunostaining of Ki-67 and (D) statistical analysis of Ki-67 expression after transfection with control or si-RhoG constructs. Data represent mean ± SD. Data were analyzed by one-way ANOVA followed by Turkey’s HSD post hoc test. **p < 0.01; ***p < 0.001.
Knockdown of RHOG altered RPE cell motility and lamellipodium formation
We next analyzed the effects of RHOG silencing on RPE cell motility and cytoskeletal structure. Monolayers of RPE cells transfected with si-RHOG had significantly reduced wound healing ability after cross-hair pattern wounding than the controls (Fig. 6A, B). In addition, transwell migration assays revealed significantly fewer si-RHOG transfected migratory cells than migratory control cells (Fig. 6C, D). We also found fewer lamellipodia and less filamentous actin condensation at the borders of RPE cells through phalloidin staining after transfection with si-RHOG (Fig. 6E). Western blot analysis after miR-124 knockdown and si-RHOG treatment revealed similar knockdowns of RHOG and RAC1 in RPE cells (Fig. 6F).
RhoG-silencing effects on RPE migration. (A) Representative images of cross-hair wound healing in monolayers of RPE cells transfected with si-RhoG or control constructs. (B) Statistical analysis of the assay in panel (A). (C) Representative images and (D) statistical analysis of a transwell migration assay with cells transfected with the control vector or si-RhoG. (E) Phalloidin staining for lamellipodium formation and filamentous actin condensation at the borders of RPE cells transfected with control or si-RhoG constructs. (F) Western blot analysis of the expression of RhoG and its downstream effector Rac1 in RPE cells transfected with miR-124 and si-RhoG. Data represent mean ± SD. Data were analyzed by independent t test. *p < 0.05; **p < 0.01.
Transfection with miR-124 and si-RHOG resulted in similar expression patterns of cell cycle regulatory factors
Transfection with miR-124 and si-RHOG resulted in knockdown of RHOG in RPE cells. In addition, miR-124 transfection induced down-regulation of the cell cycle-related factors, Cdk2, Cdk4, p21Waf1/Cip1, Cyclin D1, and Cyclin D3, in RPE cells. Conversely, p27Kip1 was up-regulated after transfection with miR-124. Knockdown of RHOG with siRNA similarly regulated the expression of cell cycle-related proteins (Fig. 7A, B).
RhoG knockdown or replenishment in the context of miR-124 regulation proved miR-124 plays a pivotal role in RhoG-mediated cell proliferation and migration
To definitely disclose the relationship between miR-124 and RhoG, we conducted a series of experiments in the context of simultaneous modulation of miR-124 and RhoG expression. Overexpression of miR-124 together with RhoG replenishment resulted in the rescue of RhoG expression in RPE cells; although RhoG expression was downregulated by miR-124 overexpression, the supplementation with rhRhoG increased the RhoG levels (expectedly), as per the western blot analysis. In addition, RhoG overexpression secondary to miR-124 inhibition was blocked by siRhoG transfection (Fig. 8A). Of note, cell viability and wound healing potential were recovered after treatment with rhRhoG (in the context of miR-124 overexpression), similarly to the observed after inhibition of endogenous miR-124 (Fig. 8B–E).
RhoG knockdown and rescue experiments in RPE cells differentially expressing miR-124. (A) Western blot analysis of RhoG expression in the context of miR-124 overexpression and RhoG overexpression, or miR-124 inhibition and siRhoG transfection. (B,C) Quantitative measurements of RPE cell viability (measured via the WST-8 assay) and the respective microscopic images in the context of miR-124 overexpression and RhoG overexpression, or miR-124 inhibition and siRhoG transfection. (D,E) Wound healing assay and the respective quantitative analysis in the context of miR-124 overexpression and RhoG overexpression, or miR-124 inhibition and siRhoG transfection. Data represent mean ± SD. Data were analyzed by one-way ANOVA followed by Turkey’s HSD post hoc test. *p < 0.05; **p < 0.01; ***p < 0.001.
Discussion
During development, RPE cells undergo mitotic cell division; however, they become terminally differentiated at the embryonic stage20. Hence, human RPE cells remain in a quiescent stage of mitosis in the postnatal period. In an animal model of RPE atrophy, the endothelium rapidly healed via proliferation and migration of neighbouring RPE cells after debridement21,22,23. However, in humans, RPE regeneration is limited and accompanied by pathological epithelial–mesenchymal transition (EMT)24. The lack of regenerative capacity in the human CNS, including the retinal system, necessitates studies on cell-based therapeutic modalities for conditions such as dry age-related macular degeneration25,26,27. Contrary to the usual lack of proliferation in the CNS, formation of subretinal membranes in proliferative vitreoretinopathy11,12, tractional membranes in diabetic retinopathy13, and idiopathic epiretinal membranes14 is induced by the abnormal proliferation and migration of RPE cells. Understanding the underlying mechanisms controlling RPE cell proliferation and migration is essential to the development of therapeutic agents for proliferative RPE diseases.
In a previous report, we elucidated the inhibitory effects of miR-124 on the TGF-β1-induced EMT of RPE cells28. Although TGF-β1 itself is a potent growth inhibitor of most cell types, we found decreased cell density after transfection of miR-124 into RPE cells29,30. Furthermore, RHOG is a known target of miR-124, and its nomenclature emphasizes its role in growth; thus, we hypothesized that miR-124-regulated RhoG would control the cell cycle progression or proliferation of RPE cells16,28,31. The expression of miR-124 dynamically changed along with RPE cell confluence, which is consistent with miRNA in situ hybridization findings in murine RPE cells, which revealed decreased expression of miR-124 upon confluence8. However, the expression of miR-124 was significantly increased in RPE cells at between 50 and 90% confluence. RhoG expression, which might be affected by increment in endogenous expression of miR-124, showed a similar, but delayed pattern to miR-124 expression. Because miRNAs mainly control the posttranscriptional regulation of their targets, a delayed decrease in RhoG, compared to miR-124, expression is logical. However, although miR-124 was significantly decreased after full confluence, RhoG expression level did not increase again; in fact, it further decreased. This was contrary to our hypothesis that miR-124 regulates RhoG expression. The activity of the Rho protein is determined by a variety of physical factors, such as integrin engagement, confluence, and formation of adherent junctions32,33. It was speculated that this pattern would be due to the difficulty to maintain low confluence continuously in an in vitro system; of note, cell-to-cell contact in the context of full confluence during long-term RPE culture has a strong inhibitory effect on cell proliferation32,33. Therefore, to demonstrate the regulation of RhoG expression by miR-124, we decided to treat cells with a miR-124 inhibitor for 24 h and compare them with non-treated cells. Importantly, after treatment with the miR-124 inhibitor, RhoG expression was elevated at 72 h compared to that in non-treated cells at the same time-point.
Various studies have reported regulation of RPE cell proliferation by specific miRNAs. Targeting of PAX6 by miR-328 led to down-regulation of Pax6 and, consequently, increased RPE cell proliferation34. Adijanto et al.35 also reported that miR-204 and -211 regulate various physiological processes, including RPE cell proliferation. Most recently, miR-34a was reported to regulate RPE cell proliferation by targeting c-Met and LGR4; the authors also reported that proliferating RPE cells had low endogenous expression of miR-34a10,36. The proliferation of various tumour cells is controlled by their endogenous expression levels of miR-124. The expression of miR-124 was low in squamous carcinoma and breast cancer cells, and the overexpression of miR-124 suppressed tumour cell proliferation and attenuated viability37,38. In those reports, miR-124 targeted SphK1 and Cdk4 in squamous carcinoma and breast cancer cells, respectively. Especially, in the eye, miR-124 is necessary for both cell proliferation and repression of neurogenesis at the optic vesicle stage in Xenopus39. In brain neural cells, high expression of miR-124 negatively regulates cell cycle progression beyond the G0/G1 phase by repression of Cdk640,41. We found that Cdk2, Cdk4, Cyclin D1, and Cyclin D3 were down-regulated by miR-124 overexpression, consistent with increased expression of p27Kip1, in RPE cells. Since these markers were regulated in the same manner by si-RHOG and we could demonstrate that miR-124 mainly targets RHOG (via RhoG knockdown and rescue experiments), we conclude that the miR-124-induced regulation of the proliferation of RPE cells is accomplished via RHOG. Therefore, miR-124 may have an additional mechanism of action in RPE cells compared to that in the neural cells of the CNS. Further research is needed to elucidate the underlying mechanisms driving the increased expression of p27Kip1 upon miR-124 overexpression in RPE cells.
Accelerated cell migration, with concurrent cell cycle progression, plays an important role during embryonic development and wound healing. High lens and RPE cell motility contribute to pathological alterations, such as posterior capsule opacification or proliferative vitreoretinopathy-related changes to the retina. The regulation of motility would be a putative therapeutic approach to repress the visual impairment caused by posterior capsule opacification or proliferative vitreoretinopathy. Cell motility is an integrated, multistep cycle, which includes cell polarisation, protrusion, stable focal adhesion, detachment of adhesion, and retraction42,43. Small GTPases in the RHO family control elements of cell motility, which led us to hypothesize that the repression of RPE cell motility by miR-124 overexpression acted via the miR-124/RHOG/RAC1 signalling axis. The binding of miR-124 to the 3′ UTR of RHOG mRNA, and its regulation of lamellipodium formation, wound healing, and RPE cell migration, are consistent with that hypothesis.
In conclusion, the miR-124/RHOG axis might be a potential therapeutic target in proliferative diseases affecting RPE cells, such as proliferative vitreoretinopathy.
Methods
Cell line and cell culture
All experiments in this study used an immortalized human RPE cell line (APRE-19), which was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). APRE-19 was maintained in a 1:1 mixture of Dulbecco’s modified Eagle’s medium and Ham’s F12 nutrient supplemented with 10% (v/v) foetal bovine serum (FBS) and 1% (v/v) penicillin/streptomycin in a humidified incubator at 37 °C with 5% CO2 (all reagents from Welgene, Daegu, Korea). The cell line was used in experiments between passages 10 and 18.
RNA interference constructs and reagents
mirVana™ miR-124 mimic (miR-124), miRNA mimic negative control (miR-NC), mirVana™ miR-124 inhibitor, miRNA inhibitor-NC, Lipofectamine® RNAiMAX and Lipofectamine™ 3,000 transfection reagent, and Alexa Fluor® 488-conjugated phalloidin were purchased from ThermoFisher Scientific (Waltham, MA, USA). The siRNA NC and 4 predesigned RHOG siRNAs (si-RHOG) with different sequences were from Qiagen (Hilden, Germany). Vectashield® Mounting Medium with DAPI was purchased from Vector Laboratories (Peterborough, UK). The 5-bromo-2′-deoxyuridine (BrdU) enzyme-linked immunosorbent assay (ELISA) kit was purchased from Roche Diagnostics (Basel, Switzerland), Cell Counting Kit-8 (CCK-8) from Dojindo Molecular Technologies (Rockville, MD, USA), and the transwell migration kit from Cell Biolabs (San Diego, CA, USA). The following primary antibodies were used: anti-RHOG from Sigma-Aldrich (St. Louis, MO, USA); anti-RAC1 from EMD Millipore (Billerica, MA, USA); anti-GAPDH, Cyclin D1, D3, Cdk2, Cdk4, p21Waf1/Cip1, and p27Kip1 from Cell Signaling Technology (Danvers, MA, USA); and anti-Ki-67 from Merck Millipore (Billerica, MA, USA) and Abcam (Cambridge, UK). All secondary antibodies for immunoblot assays were from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and Jackson Laboratory (Bar Harbor, ME, USA), and fluorescently conjugated secondary antibodies for immunostaining were from Molecular Probes (Eugene, OR, USA). All other chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA).
Quantitative analysis of relative miR-124 expression
To investigate the relative expression levels of endogenous miR-124 depending on RPE cell density, ARPE-19 cells were plated at a density of 7.5 × 104 cells/mL in 6-well cell culture plates. At 24, 48, 72, and 96 h after plating, miRNA was extracted and miRNA real-time quantitative PCR (qPCR) analysis was performed. At each time point, proteins were also extracted for the detection of relative RHOG expression. The culture medium was aspirated and cells were rinsed thrice with phosphate-buffered saline (PBS); thereafter, total RNA was extracted using QIAzol lysis reagent (Qiagen, Hilden, Germany). A 1-μg aliquot of total RNA was reverse transcribed using miScript II Reverse Transcriptase (Qiagen, Hilden, Germany). qPCR was performed using QuantiTect® SYBR® Green PCR Master Mix and the Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). The expression of miR-124 was normalised to that of the control RNU6 RNA or miR-16 using the 2−ΔΔCT method17.
Cell viability and proliferation assays
ARPE-19 cells were plated in 96-well plates and subsequently transfected with oligonucleotides (miRNA mimic or siRNA) after 24 h. We introduced 50 nM miR-NC or miR-124 into the cells with RNAiMAX transfection reagent over 6 h, and 50 nM of si-NC or si-RHOG molecules were transfected using the same reagent. Twenty-four hours after transfection, cell viability or BrdU incorporation assays were performed. Cell viability was evaluated with a CCK-8 assay. Briefly, the culture medium was aspirated and cells were rinsed thrice with PBS. A mixture of 10 µL reagent with 90 µL serum-free medium was added to each well of a 96-well plate. After a 4-h incubation, colorimetric detection was performed at 450 nm with a microplate reader (VersaMax, Molecular Devices, Sunnyvale, CA, USA). Proliferation was assessed with a BrdU ELISA kit. Briefly, BrdU labelling reagent was added to the culture medium and incubated for 2 h prior to the assay. Culture medium was aspirated after 2 h, followed by fixation, denaturation, and anti-BrdU peroxidase conjugation. The substrates were added to each well and colorimetric detection was performed with a microplate reader. All experiments were performed in triplicate.
Western immunoblot analyses
Cells were rinsed thrice with cold PBS and lysed in radioimmunoprecipitate buffer. The protein concentrations of the lysates were measured using a protein bicinchoninic acid assay kit (ThermoFisher Scientific, Waltham, MA, USA). Equal quantities of total protein samples were loaded in 8–15% polyacrylamide gels for sodium dodecyl sulphate polyacrylamide gel electrophoresis. The resolved proteins were electroblotted onto nitrocellulose membranes and blocked with 5% skim milk solution for 1 h. After overnight incubation with the appropriate primary antibodies, the membranes were incubated with secondary antibodies for 1 h. GAPDH was used as a loading control for blot band densitometry.
Phalloidin staining and immunocytochemistry
ARPE-19 cells were plated on 4-well cell culture slides (SPL Life Sciences, Pocheon, Korea). The miRNA or siRNA constructs were delivered 24 h after plating and the cells were cultured to 50–60% confluency. The cells were rinsed thrice with PBS and fixed with 4% paraformaldehyde for 30 min. For observation of filamentous actin, the cells were stained with Alexa Fluor® 488-conjugated phalloidin and Hoechst 33,342 solution. For immunocytochemical analyses, cells were permeabilised with 0.4% (v/v) Triton™ X-100 for 15 min and blocked with 1% (w/v) bovine serum albumin in 0.1% (v/v) Tween® 20 in PBS for 30 min. They were incubated with anti-Ki-67 primary antibodies for 12 h, then Alexa Fluor® 594-conjugated secondary antibodies (Molecular Probes, Eugene, OR, USA) for 2 h at room temperature. RPE cell nuclei were stained with Vectashield® mounting solution containing 4′,6-diamidino-2-phenylindole (DAPI) after several rinses. Each slide was observed using an upright fluorescent microscope (Axio Imager 2; Zeiss, Cologne, Germany) and images were obtained using AxioVision SE64 version 4.9.1.0 software (Zeiss, Cologne, Germany). Ki-67-positive cells were counted at 5 different locations, and the percentage of Ki-67-positive cells of DAPI-positive cells was calculated. All experiments were performed in triplicate.
Wound-healing assay
APRE-19 cells were plated into 12-well plates and allowed to reach 100% confluence. After cell transfection with RNA interference constructs (miRNA mimic, miRNA inhibitor, or siRNA), a cross-hair linear wound was inflicted with a 200-µL pipette tip followed by treatment with 50 μg/ml mitomycin C for 1 h. After 3 PBS rinses, medium containing 1% FBS was added. The remaining wound was observed at each indicated time point and measured under phase-contrast microscopy using ImageJ (https://imagej.nih.gov/ij/; provided in the public domain by the National Institutes of Health, Bethesda, MD, USA). All experiments were performed in triplicate.
Transwell migration assay
After transfection with miRNA or siRNA, cells were trypsinised and resuspended in serum-free medium. Medium containing 10% FBS was added to the bottom chambers of the wells and cell suspensions were poured into polycarbonate membrane inserts with 8-µm pores. After a 12-h incubation, the cells that had migrated to the bottom of the polycarbonate membrane were fixed with 4% paraformaldehyde and stained with haematoxylin. Migratory cells were counted with a phase-contrast microscope at 3 different locations. All experiments were performed in triplicate.
Plasmid transfection and luciferase reporter assay
A random control luciferase plasmid and LightSwitch™ RHOG mRNA 3′ untranslated region (UTR) plasmid were purchased from SwitchGear Genomics (Menlo Park, CA, USA). Cells were plated in 96-well luminescence plates and conventional 96-well cell culture plates prior to transfection and cultured up to 70% confluence over 24 h. The cells were co-transfected for 6 h, with either random control or RHOG mRNA 3′ UTR luciferase plasmid and either miR-124 or miR-NC, using Lipofectamine™ 3,000 reagents, then incubated for 24 h. The transfected cells were lysed with buffer-substrate solution. After 30 min of incubation, luciferase activity of each well was detected with a luminometer (Centro LB 960; Berthold Technologies, Bad Wildbad, Germany). All experiments were performed in triplicate.
Statistical analysis
Representative data obtained from experiments conducted in triplicate are presented as mean ± standard deviation. Data were evaluated using independent t tests, or one-way analysis of variance (ANOVA) and Tukey’s honest significant difference post hoc test, to identify statistically significant differences. A p value < 0.05 was regarded as statistically significant.
References
Etienne-Manneville, S. & Hall, A. Rho GTPases in cell biology. Nature 420, 629–635 (2002).
Vincent, S., Jeanteur, P. & Fort, P. Growth-regulated expression of rhoG, a new member of the ras homolog gene family. Mol. Cell Biol. 12, 3138–3148 (1992).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Macfarlane, L. A. & Murphy, P. R. MicroRNA: biogenesis, function and role in cancer. Curr. Genom. 11, 537–561 (2010).
Lagos-Quintana, M. et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739 (2002).
Sempere, L. F. et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13 (2004).
Karali, M. et al. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genom. 11, 715 (2010).
Karali, M., Peluso, I., Marigo, V. & Banfi, S. Identification and characterization of microRNAs expressed in the mouse eye. Invest. Ophthalmol. Vis. Sci. 48, 509–515 (2007).
He, S. et al. Soluble EphB4 inhibition of PDGF-induced RPE migration in vitro. Invest. Ophthalmol. Vis. Sci. 51, 543–552 (2010).
Hou, Q. et al. Inhibitory effect of microRNA-34a on retinal pigment epithelial cell proliferation and migration. Invest. Ophthalmol. Vis. Sci. 54, 6481–6488 (2013).
Pastor, J. C., de la Rua, E. R. & Martin, F. Proliferative vitreoretinopathy: risk factors and pathobiology. Prog. Retin. Eye Res. 21, 127–144 (2002).
Tosi, G. M., Marigliani, D., Romeo, N. & Toti, P. Disease pathways in proliferative vitreoretinopathy: an ongoing challenge. J. Cell. Physiol. 229, 1577–1583 (2014).
Hiscott, P., Gray, R., Grierson, I. & Gregor, Z. Cytokeratin-containing cells in proliferative diabetic retinopathy membranes. Br. J. Ophthalmol. 78, 219–222 (1994).
Smiddy, W. E. et al. Idiopathic epiretinal membranes ultrastructural characteristics and clinicopathologic correlation. Ophthalmology 96, 811–820 (1989) (discussion 821).
Ivan, M. & Huang, X. miR-210: fine-tuning the hypoxic response. Adv. Exp. Med. Biol. 772, 205–227 (2014).
Franke, K. et al. miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 31, 2908–2921 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Friedman, R. C., Farh, K. K., Burge, C. B. & Bartel, D. P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 (2009).
Grimson, A. et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 27, 91–105 (2007).
Fuhrmann, S., Zou, C. & Levine, E. M. Retinal pigment epithelium development, plasticity, and tissue homeostasis. Exp. Eye Res. 123, 141–150 (2014).
Del Priore, L. V. et al. Debridement of the pig retinal pigment epithelium in vivo. Arch. Ophthalmol. 113, 939–944 (1995).
Del Priore, L. V., Kaplan, H. J., Hornbeck, R., Jones, Z. & Swinn, M. Retinal pigment epithelial debridement as a model for the pathogenesis and treatment of macular degeneration. Am. J. Ophthalmol. 122, 629–643 (1996).
Lopez, P. F. et al. Retinal pigment epithelial wound healing in vivo. Arch. Ophthalmol. 113, 1437–1446 (1995).
Tamiya, S., Liu, L. & Kaplan, H. J. Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Invest. Ophthalmol. Vis. Sci. 51, 2755–2763 (2010).
Buchholz, D. E. et al. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl. Med. 2, 384–393 (2013).
Schwartz, S. D. et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 379, 713–720 (2012).
Schwartz, S. D. et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet 385, 509–516 (2015).
Jun, J. H. & Joo, C. K. MicroRNA-124 controls transforming growth factor beta1-induced epithelial-mesenchymal transition in the retinal pigment epithelium by targeting RHOG. Invest. Ophthalmol. Vis. Sci. 57, 12–22 (2016).
Huang, S. S. & Huang, J. S. TGF-beta control of cell proliferation. J. Cell. Biochem. 96, 447–462 (2005).
Moses, H. L. TGF-beta regulation of epithelial cell proliferation. Mol. Reprod. Dev. 32, 179–184 (1992).
Schumacher, S. & Franke, K. miR-124-regulated RhoG: a conductor of neuronal process complexity. Small GTPases 4, 42–46 (2013).
Ribatti, D. A revisited concept: contact inhibition of growth. From cell biology to malignancy. Ribatti D. Exp Cell Res. 359, 17–19 (2017).
Kim, N., Koh, E., Chen, X. & Gumbiner, B. M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci USA 108, 11930–11935 (2011).
Chen, K. C. et al. MicroRNA-328 may influence myopia development by mediating the PAX6 gene. Invest. Ophthalmol. Vis. Sci. 53, 2732–2739 (2012).
Adijanto, J. et al. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J. Biol. Chem. 287, 20491–20503 (2012).
Hou, Q. et al. LGR4 is a direct target of microRNA-34a and modulates the proliferation and migration of retinal pigment epithelial ARPE-19 cells. PLoS ONE 11, e0168320 (2016).
Feng, T. et al. MiR-124 inhibits cell proliferation in breast cancer through downregulation of CDK4. Tumour Biol. 36, 5987–5997 (2015).
Zhao, Y. et al. MiR-124 acts as a tumor suppressor by inhibiting the expression of sphingosine kinase 1 and its downstream signaling in head and neck squamous cell carcinoma. Oncotarget 8, 25005–25020 (2017).
Liu, K. et al. MiR-124 regulates early neurogenesis in the optic vesicle and forebrain, targeting NeuroD1. Nucleic Acids Res. 39, 2869–2879 (2011).
Cheng, L. C., Pastrana, E., Tavazoie, M. & Doetsch, F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 (2009).
Silber, J. et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6, 14 (2008).
Katoh, H., Hiramoto, K. & Negishi, M. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 119, 56–65 (2006).
Katoh, H. & Negishi, M. RhoG activates Rac1 by direct interaction with the Dock180-binding protein Elmo. Nature 424, 461–464 (2003).
Acknowledgements
The authors thank Jun-Sub Choi, PhD, and Yong-Soo Byun, MD, PhD, Catholic Institute for Visual Science, The Catholic University of Korea, College of Medicine, Seoul, Korea, and Youngkyun Lee, PhD, Department of Oral Biochemistry, School of Dentistry, IHBR, Kyungpook National University, Daegu, Korea, for advice on experiments.
Funding
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea Government (Ministry of Science, ICT & Future Planning; Nos. 2014R1A5A2010008 and NRF-2015R1C1A1A02037062).
Author information
Authors and Affiliations
Contributions
Conceptualization: J.H.J., H.-G.L., C.-K.J. Data curation: J.H.J., H.-G.L. Formal analysis: J.H.J., M.-J.S., H.-G.L., W.-K.B., J.-Y.K. Funding acquisition: J.H.J., C.-K.J. Investigation: J.H.J., M.-J.S., H.-G.L. Methodology: J.H.J., H.-G.L. Project administration: J.H.J. Resources: C.-K.J. Software: J.H.J. Supervision: C.-K.J. Validation: J.H.J., H.-G.L. Visualization: J.H.J. Writing—original draft: J.H.J. Writing—review and editing: J.H.J., K.Y.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jun, J.H., Son, MJ., Lee, HG. et al. Regulation of Ras homolog family member G by microRNA-124 regulates proliferation and migration of human retinal pigment epithelial cells. Sci Rep 10, 15420 (2020). https://doi.org/10.1038/s41598-020-72360-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-72360-5
This article is cited by
-
MicroRNA-124 conducts neuroprotective effect via inhibiting AK4/ATF3 after subarachnoid hemorrhage
Experimental Brain Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.