Abstract
Genetic polymorphisms of NEIL1 and NEIL2 maybe change protein function, and increased carcinogenesis. In this study, seven NEIL1 SNPs and three NEIL2 SNPs were selected. 400 CSCCs, 400 CIN III, and 1200 normal healthy controls were genotyped by mismatch amplification PCR. mRNA and protein expression of NEIL2 was measured in 92 freshly-obtained CSCC tumor tissues. The association between homozygote CC genotype of NEIL2 rs804270 with susceptible risk was gradually increased in CIN III (OR = 1.44) and CSCC (OR = 2.22). Carriers of C-allele (GC + CC) at rs804270 had a high risk of CSCC (OR = 1.46). The heterozygote GT genotype of rs8191664 was also closely related to the higher risk of CINIII (OR = 1.59) and CSCC (OR = 2.54). Carriers of T-allele (GT + TT) at rs8191664 had a high risk for CIN III (OR = 1.55) and CSCC (OR = 2.34). The genotypes of NEIL2 rs804270 (G/C) and rs8191664 (G/T) that were related to the higher risk for CIN III were CC-GG (OR = 1.42) and CC-GT (OR = 2.07). More notably, there was a greater risk for CSCC with the GC-GT (OR = 1.91), CC-GG (OR = 1.67), and CC-GT (OR = 6.18) genotypes. NEIL2 mRNA expression in CSCCs with the rs804270-CC genotype was lower expression than those in CSCCs with the rs804270-GG and rs804270-GC genotypes. Similarly, NEIL2 protein expression was significantly decreased in CSCCs with the rs804270-CC genotype. In summary, the two genetic polymorphisms (rs804270 and rs8191664) of NEIL2 gene were significantly associated to the increased susceptibility of CIN III or CSCC. This increased susceptibility maybe due to altered NEIL2 repair activity through altered protein expression, or changed structure of the functional domain. The genotypes of GC-GT, CC-GG, and CC-GT of rs804270 and rs8191664 of NEIL2 gene could act as a genetic predictive biomarker of susceptibility to CIN III and CSCC.
Similar content being viewed by others
Introduction
Globally, cervical carcinoma is the fourth most common malignant cancer in women with approximately 500,000 new cases and almost 300,000 deaths each year1. Cervical carcinoma is also the third leading cause of cancer-related death in women; this is very worrying because the incidence of this disease is rising2. The primary etiological factor is the infection by the high risk human papilloma virus (HR-HPV)3. Aside from breast cancer, cervical carcinoma has become the most common form of cancer in Chinese women (with an incidence of 98.9 patients per 100,000 of the Chinese population). The mortality rate from cervical carcinoma has increased to 30.5 per 100,0004. However, while 80% of women will become infected with HPV during their lifetime, only a small number will develop malignant cervical carcinoma5. Epidemiological evidence has confirmed that a range of genetic variations are associated with the risk of cervical carcinoma6.
Previous research, including two genome-wide studies, have identified loci that are genetically susceptible and genetic polymorphisms that are closely related to the occurrence of cervical carcinoma6,7,8. However, these genetic polymorphisms account for only a small part of the genetic susceptibility to cervical cancer. Therefore, more comprehensive and in-depth genetic study is needed to further understand the genetic risk factors for cervical carcinoma.
The integrity and genetic stability of the genome are maintained by a variety of DNA repair systems in order to combat environmental attacks, replication mistakes and cumulative geriatric degeneration. There are five major DNA repair mechanisms in the human genome that are used to repair damaged DNA, including direct reversal, nucleotide excision repair, base excision repair, mismatch repair and recombination repair; previous studies have shown that more than 100 genes are involved in these mechanisms9.
During mammalian cell replication, the repair of damaged DNA caused by reactive oxygen species (ROS) is mainly performed by a group of DNA glycosylases, including two DNA glycosylase genes, NEIL1 and NEIL2. NEIL1 and NEIL2 can protect normal somatic cells from radiation damage; if these genes are subject to functional genetic variation, then it is likely that their normal protein function may be changed, eventually leading to a change in cell fate and increased carcinogenic potential10,11,12,13. Several reports have shown that genetic variations in NEIL1 and NEIL2 are significantly associated with susceptibility to solid malignant tumors, such as oropharyngeal cancer14, gastric cancer15, bladder cancer16 and colorectal adenoma17. However, from these published studies, we found that the association analysis data of single nucleotide polymorphisms (SNPs) of these two genes with cancer risk are not comprehensive, and their protein expression and functional activity have not been generally studied. In addition, the correlation between SNP loci in NEIL1 and NEIL2 and susceptibility to cervical carcinoma has not been studied so far.
Therefore, in our large-sample population-based study, we selected seven SNP loci in NEIL1 and three SNP loci in NEIL2, then investigated their genotype frequency in 400 cervical squamous cell carcinomas (CSCC), 400 cervical intraepithelial neoplasias (CIN III) and 1200 normal healthy controls, and analyzed the association between these SNPs in the NEIL1 and NEIL2 genes and susceptibility to CSCC and CIN III. Furthermore, we also detected the expression of the NEIL2 gene in different genotypes of cervical cancer cells at the mRNA and protein level to investigate the relationship between SNP genotypes and gene expression. The purpose of this study was to better understand the potential role of specific SNP genotypes in the carcinogenesis of CSCC.
Results
The relationship between genetic polymorphisms in NEIL1 and NEIL2 and the risk of CIN III or CSCC
As show in Table 1, the genotype or allele frequencies of genetic polymorphisms in NEIL1 and NEIL2 were rs4462560, rs7182283, rs7402844, rs5745920, rs8030014, rs11634109 and rs79244935 for NEIL1, and rs804270, rs8191613 and rs8191664 for NEIL2. Hardy-Weinberg Equilibrium(HWE) test was performed for all of SNP alleles in normal healthy control group(Shown in Table S2), the P value of HWE analysis of some loci is less than 0.05, which indicates that the specific genotypes of these loci have certain enrichment in Chinese population. Combined with the statistical results of Tables 1 and 2, we believe that the enrichment of some loci in normal healthy control group does not affect the comparison of genotype frequency between disease group and normal healthy control group.
The frequency of genotype identified that all seven of the NEIL1 genetic polymorphisms (rs4462560, rs7182283, rs7402844, rs5745920, rs8030014, rs11634109 and rs79244935) and the NEIL2 rs8191613 genetic polymorphism were not associated with the risk of CIN III and CSCC. The GG, GC, and CC genotype frequencies of NEIL2 rs804270 were 30.7%, 48.8% and 20.5% in normal healthy controls; 28.3%, 44.5% and 27.3% in CIN III and 23.3%, 42.3% and 34.5% in CSCC, respectively. These results showed that patients with the rs804270 homozygous CC genotype had a significantly higher risk of CIN III (odds ratio[OR] = 1.44; 95% confidence interval[CI]:1.06–1.97) and CSCC (OR = 2.22; 95%CI: 1.63–3.02). We also found that the frequency of C alleles at the rs804270 locus in CIN III (396/800, 49.5%) and CSCC (445/800, 55.6%) were significantly higher than those in normal healthy controls (1078/2400, 44.9%). The OR of the C allele in CIN III was 1.20 (95%CI: 1.02–1.41) and 1.54 (95%CI: 1.31–1.81) in CSCC. Carriers of the C-allele (GC + CC) at rs804270 were associated with a higher risk for CSCC (OR = 1.46; 95%CI: 1.12–1.90).
The GG, GT and TT genotype frequencies of NEIL2 rs8191664 were 85.9%, 11.8% and 2.3% in the normal healthy controls; 79.8%, 17.5% and 2.8% in CIN III and 72.3%, 25.3% and 2.5% in CSCC, respectively. These results showed that women carrying the heterozygote GT genotype rs8191664 also had a significantly elevated risk of CIN III (OR = 1.59; 95%CI: 1.17–2.18, P = 0.003) and CSCC (OR = 2.54; 95%CI: 1.91–3.38, P = 0.0001). The T allele frequencies of rs8191664 in CIN III (92/800, 11.5%) and CSCC (121/800, 15.1%) were higher than those in normal healthy controls (196/2400, 8.2%). The T allele was associated with a higher risk for both CIN III (OR = 1.46; 95%CI: 1.23–1.90) and CSCC (OR = 2.00; 95%CI: 1.57–2.55), respectively. Carriers of the T-allele (GT + TT) at rs8191664 were associated with a higher risk for CIN III (OR = 1.55; 95%CI: 1.16–2.08) and CSCC (OR = 2.34; 95%CI: 1.78–3.08).
False discovery rate (FDR) multiple testing corrections were applied in order to avoid Type I errors. We found that the frequency of CC or GC + CC genotype of rs804270 and GT or GT + TT genotype of rs8191664 in CSCC group were still higher than normal healthy control group. The specific statistics are shown in the Pa value in Table 1.
The relationship between genetic polymorphisms in NEIL1 and NEIL2 and HR-HPV-positive cases of CIN III and CSCC
In the HR-HPV-positive groups, NEIL1 rs4462560, rs7182283, rs7402844, rs5745920, rs8030014, rs11634109 and rs79244935, and NEIL2 rs8191613 genetic polymorphisms were not related to the risk of CIN III or CSCC (Table 2).
However, the homozygous CC genotype of rs804270 showed relatively higher risk for CIN III (OR = 1.80; 95%CI: 1.08–2.97) and CSCC (OR = 2.36; 95%CI: 1.33–4.17). The elevated risk of CIN III and CSCC with the C allele showed an OR of 1.36 (95%CI: 1.05–1.76) and 1.59 (95%CI: 1.19–2.13), respectively. For rs8191664, the heterozygous GT allele also showed a relatively higher risk of CIN III (OR = 2.03; 95%CI: 1.22–3.36) and CSCC (OR = 2.82; 95%CI: 1.65–4.84) in the HR-HPV-positive group. The increased risk of the T allele for CIN III and CSCC showed an OR of 1.60 (95%CI: 1.05–2.44) and 2.12 (95%CI: 1.35–3.31), respectively. Carriers of the T-allele (GT + TT) at rs8191664 were associated with a higher risk for CIN III (OR = 1.85; 95%CI: 1.15–2.96) and CSCC (OR = 2.56; 95%CI: 1.55–4.25).
After FDR multiple testing corrections, we also found that the frequency of CC genotype of rs804270 and GT or GT + TT genotype of rs8191664 in CSCC group were still higher than normal healthy control group. The specific statistics are shown in the Pa value in Table 2.
The association between NEIL2 rs804270 and rs8191664 genetic polymorphisms and sexual and reproductive histories in patients with CIN III and CSCC
Stratified analysis was performed to analyze the association between the NEIL2 rs804270 and rs8191664 genotypes and age, age at first intercourse, number of sexual partners, number of parities, HR-HPV infection and other clinical data. There was no enrichment between subgroups with CIN III and CSCC and the NEIL2 rs804270 genetic polymorphism, as show in Table 3. However, as show in Table 4, we observed a higher enrichment of the NEIL2 rs8191664 genetic polymorphism when patients were subgrouped by the number of sexual partners in CIN III (χ2 = 15.577, P = 0.0001) and CSCC (χ2 = 26.556, P = 0.0001).
Association analysis between the NEIL2 rs804270 (G/C) and rs8191664 (G/T) genotypes and the risk of CINIII and CSCC
We analyzed the genotype linkage pattern between the frequencies of both rs804270(G/C) and rs8191664(G/T) genotypes because there was a significant association between these two genetic polymorphisms with the risk of CINIII and CSCC. As shown in Table 5, the GG-TT and CC-TT genotypes were not detected in any of the cases and normal healthy controls. Compared with the reference genotype GG-GG, the CC-GG (OR = 1.42; 95%CI: 1.01–2.00) and CC-GT (OR = 2.07; 95%CI: 1.19–3.61) genotypes were significantly associated with an increased risk of CIN III. A higher risk was detected for GC-GT (OR = 1.91; 95%CI: 1.13–3.23), CC-GG (OR = 1.67; 95%CI: 1.16–2.37) and CC-GT (OR = 6.18; 95%CI: 3.85–9.93) in CSCCs. These data indicated that the genotype linkage pattern of the CC homozygous genotype of rs804270(G/C), and the GT heterozygous genotype of rs8191664(G/T), was associated with an elevated risk for CIN III and CSCC.
In addition, the CC-GG genotype was the most common genotype linkage pattern in the CIN III [85/(85 + 24), 77.98%], CSCC [80/(80 + 58), 57.97%] and nornal healthy control groups [206/(206 + 40), 83.74%] which carried the CC genotype at rs804270(G/C). Similarly, the GC-GT genotype was the most common genotype linkage pattern in the nornal healthy control [58/(44 + 58 + 40), 40.85%] and the CIN III [27/(19 + 27 + 24), 38.57%] groups, which carried the GT genotype at rs8191664 (G/T). However, the CC-GT genotype was the most common genotype linkage pattern in the CSCC group [58/(17 + 26 + 58), 57.43%] which carried the GT genotype at rs8191664 (G/T). These results indicate that there was a specific genotype linkage pattern between rs804270(CC) and rs8191664(GT). In other words, these specific genotype linkage patterns were associated with a higher risk of CIN III or CSCC. The genotypes of GC-GT, CC-GG, and CC-GT of rs804270 and rs8191664 SNP in the NEIL2 gene may act as a genetic predictive biomarker of susceptibility for CIN III and CSCC.
The linkage disequilibrium and haplotype analysis of three SNP loci in NEIL2 gene
Because the genotypes of two SNP loci in NEIL2 were significantly correlated with the susceptibility of CIN III and CSCC, we further analyzed the linkage disequilibrium and haplotype of all three SNP loci in NEIL2 with the SHEsis software. The pairing analysis showed that the D’ and r2 values did not have statistical significance, there was no linkage disequilibrium between the three SNPs each other, this also meant that there is no specific haplotype between the three SNP. However, we noted that there may be a trend of linkage disequilibrium between rs8191613 and rs8191664 in CIN III group(D’ = 0.768), while in CSSS group, there may be a trend of linkage disequilibrium between rs804270and rs8191664(D’ = 0.344). The specific statistical results are shown in Tables 6 and 7.
The mRNA and protein expression levels of NEIL2 in CSCC tissues with different rs804270 (G/C) or rs8191664 (G/T) genotypes
The number of cases and the frequencies of the GG, GC, and CC genotypes of rs804270 among the 92 CSCC patients were 22 (23.9%), 38 (41.3%), and 32 (34.8%) cases, respectively. When the rs804270(GG) group was used as a control group, the expression of NEIL2 mRNA in patients with rs804270(CC) (0.824 ± 0.201) was significantly lower(30% reduction, P < 0.001) than that in patients with rs804270(GG) (1.215 ± 0.213) and rs804270(GC) (1.003 ± 0.188) (Fig. 1). Similarly, in the rs804270(CC) group, the protein expression of NEIL2 also was significantly lower (50% reduction, P < 0.001) (Fig. 2A,C).
mRNA expression of NEIL2 in CSCCs with different genetic polymorphisms. rs804270-GG: rs804270 genotype is GG; rs804270-GC: rs804270 genotype is GC; rs804270-CC: rs804270 genotype is CC; rs8191664-GG: rs8191664 genotype is GG; rs8191664-GT: rs8191664 genotype is GT; rs8191664-TT: rs8191664 genotype is TT. The rs804270-GG and rs8191664-GG genotypes were used as the control groups of mRNA expression in different genotypes of rs804270 and rs8191664, respectively.
Protein expression of NEIL2 in CSCCs with different genetic polymorphisms. (A,B) Immunoblot, the molecular weight of NEIL2 and GAPDH protein is 37 kDa and 36 kDa respectively; (C,D) Analysis of protein relative expression of different genotypes. In rs804270 SNP, GC compared with GG, t = 1.819, P = 0.074; CC compared with GG, t = 16.789, P = 0.000; Compared with GC, t = 12.909, P = 0.000. In rs8191664 SNP, GT compared with GG, t = 0.437, P = 0.663; T T compared with GG, t = −0.539, P = 0.592; T T compared with GT, t = −0.511, P = 0.614.
The number of cases and the frequencies of the GG, GT and TT genotypes of rs8191664 among the 92 CSCC patients, were 63 (68.5%), 26 (28.3%), and 3 (3.2%) cases, respectively. When the rs8191664(GG) group was used as a control group, there was no significant difference in the expression of NEIL2 mRNA among patients with rs8191664(GG) (0.985 ± 0.321), rs8191664(GT) (1.103 ± 0.244) and rs8191664(TT) (0.964 ± 0.235) (Fig. 1). Similarly, there was no significant difference in the expression of NEIL2 protein when compared among different genotype groups (Fig. 2B,D).
Discussion
Aerobic respiration can produce ROS via a range of pathological processes18,19. These chemicals or free radicals can cause DNA damage20, which lead to genomic instability and eventually lead to the initiation and development of malignant tumors21,22,23,24. Most of the damaged bases are removed and repaired by DNA glycosylase and the base excision repair system (BER)25,26,27,28. NEIL1 and NEIL2 are key functional proteins in the BER pathway.
The NEIL1 gene participates in the first step of the BER repair mechanism29. It was reported that FapyA or 5S-6R thymidine glycol cannot be excised by neutral trehalase 1 (NTH1) or 8-oxoguanine glycosylase (OGG1), but can be repaired by NEIL1. However, embryonic stem cells lacking NEIL1 expression were approximately twice as sensitive to low-level radiation-induced damage as normal cells30. Studies have also shown that NEIL1 protein is more efficient than 8-oxoG in the removal of thymidine glycol and 5-hydroxyuracil from damaged DNA31,32. However, NEIL1-knockout mice developed metabolic disorder syndrome, characterized by severe obesity, dyslipidemia and fatty liver33.
Three NEIL1 promoter genetic polymorphisms (c.-3769C > T, c.-3170T > G and c.-2681TA) were found to play an important role in the development of gastric cancer15. Zhai et al. found that the NEIL2 rs804270(CC) allele was associated with the advanced stage of oropharyngeal and oral squamous cell carcinoma. However, these authors did not find any risk associated with the NEIL1 rs4462560 and rs7182283 genetic polymorphisms14. In present study, we chose seven SNPs with a MAF value of more than 5% in the NEIL1 gene and found that none of these SNPs were associated with susceptibility to CSCC or its precancerous lesion CIN III. Our results show that genetic polymorphisms in the introns of NEIL1 were not related to the occurrence of cervical carcinoma. However, further studies are now required to investigate the relationship between genetic polymorphisms in the promoter region of NEIL1 and the risk of cervical carcinoma. It is possible that genetic polymorphisms in the promoter region may alter the protein expression of the NEIL1 gene, thereby altering cell behavior. However, because the three SNPs in the NEIL1 promotor region have small MAF in the general population, it is necessary to carry out additional studies featuring a larger sample size to study this association more robustly.
NEIL2 exhibits the strongest activity for 5-hydroxyuracil and weakest activity for 5-hydroxycytosine, 8-oxoG, thymine glycol and dihydrouracil34. Low expression levels of NEIL2 may cause somatic cell DNA mutation and copy number variation, thus leading to genomic instability, oncogene activation and inhibition the expression of tumor suppressor genes35,36. Elingarami et al. evaluated the potential association between NEIL2 SNPs (rs804270, 5′-UTR promoter region) and susceptibility to gastric carcinoma, and assessed whether genotypes affected the expression of NEIL2 mRNA37, they reported that there is an increased risk of gastric cancer in patients with genetic variants of NEIL2 SNP(rs804270). Moreover, studies showed that the expression of NEIL2 mRNA was significantly different when compared across different NEIL2 genotypes. In present study, we found that the frequencies of the GG, GC, and CC genotypes of NEIL2 rs804270 were 30.7%, 48.8% and 20.5% in the normal healthy controls, 28.3%, 44.5% and 27.3% in CIN III and 23.3%, 42.3% and 34.5% in CSCC, respectively. Furthermore, there was a significant correlation between the CC homozygote of rs804270 and the risk of CIN III and CSCC. Carriers of the C-allele (GC + CC) at rs804270 were associated with a higher risk for CSCC. Considering that NEIL2 rs804270 is located in the 5′-UTR promotor region, we considered that genetic variation might affect the expression of the NEIL2 gene; we therefore measured the NEIL2 expression of the mRNA and protein. Finally, we concluded that the mRNA and protein expression of NEIL2 in pathological tissues with the genotype CC of NEIL2 SNP (rs804270) were significantly reduced. These results indicated that the effect of the NEIL2 SNP (rs804270) on the susceptibility to cervical carcinoma may be caused by alterating the expression of NEIL2, and resulting in a subsequent decline in repair to the damaged genome, thus causing genomic instability and tumor initiation.
In this study, we also evaluated the association between genetic polymorphisms in the exonic regions of NEIL2 and the risk of CSCC. The heterozygous GT genotype of NEIL2 rs8191664 was associated with an elevated risk of both CIN III and CSCC. Carriers of the T-allele (GT + TT) at rs8191664 showed a higher risk for CIN III and CSCC. Interestingly, although the GT heterozygous genotype at the rs8191664 locus was identified as a high risk factor, the TT homozygous genotype was not susceptible to disease. This may be due to the fact that there was a low incidence of the TT homozygous genotype in the population. Only 2.3%, 2.8% and 2.5% of the normal healthy control, CIN III and CSCC were identified in present study, thus resulted in fluctuations in the statistical significance.
We also found that the mRNA and protein expression of NEIL2 did not differ significantly between any genotypes of NEIL2 rs8191664. We postulate that the NEIL2 rs8191664 (R257L) SNP does not change NEIL2 expression, but instead, results in a non-synonymous change in amino acid sequence. This may result in the change of the spatial structure of protein functional domains, thus affecting functional activity. Dy et al. found that compared with wild-type cells, the level of endogenous DNA damage in cells featuring the NEIL2 variant rs8191664 (G/T; R257L) was increased38. The reduced levels of DNA repair activity in cells featuring the NEIL2 rs8191664 (R257L) missense mutation can induce genomic instability that ultimately leads to the initiation of cervical carcinoma.
In present study, as shown in Tables 3 and 4, we further stratified the clinical data relating to patient age, age at first sexual intercourse, the number of parities and age at first parity. We found that there were no associations between these features and either of the two NEIL2 SNPs [rs804270 and rs8191664 (R257L)]. These results also indicated that there was no correlation between the two NEIL2 SNPs [rs804270 and rs8191664 (R257L)] and HR-HPV infection. However, there was a higher enrichment of the NEIL2 rs8191664 GT or TT genotypes in CIN III and CSCC when there was more than one sexual partner. In a family and twin studies, Sanders AR et al. found a significant association between different sexual orientations and SNPs on chromosomes 8, 13, 14 and X39. Furtherly, Pearce E et al. found that SNP in oxytocin and dopamine receptor gene was closely related to a person’s sexual attitudes and behavior, which confirmed the relationship between social behavior with the neurochemical differences caused by SNP in human gene40. This provides a theoretical basis for understanding the correlation between SNP and behavior at the molecular biological level. Because the relationship between behavior and gene is more complex than that between tumor and gene, it is related to more gene information. The study of the relationship between phenotype and gene involves more genes or loci. In order to better identify this correlation, we believe that not only the sample size of the study needs to be increased, but also the related polymorphism sites need to be increased. We’d better do further research on genome-wide association and gene function studies.
We compared the NEIL2 rs804270 (G/C) and rs8191664 (G/T) genotypes with the reference genotype GG-GG and found that the CC-GG and CC-GT genotypes were significantly associated with an increased risk of CIN III. For CSCC, the risk was much greater for the GC-GT, CC-GG and CC-GT genotypes. In particular, the CC-GT genotype has a greater impact on disease susceptibility than when these two loci were analyzed separately, the OR values for CINIII and CSCC were 2.07 and 6.18, respectively. A higher OR suggested a synergistic effect between these two genetic polymorphisms in the NEIL2 gene. It is possible that this synergistic effect promoted the development of CIN III to eventually lead to cervical carcinoma. We also observed that neither the GG genotype nor G allele conferred the risk of disease when rs8191664 was analyzed separately, although the CC-GG genotype was still at risk. This may be because the CC genotype at rs804270 had a greater impact on disease susceptibility, while rs8191664 was not a protective factor. The effect of the CC genotype at the rs804270 locus could not be eliminated by rs8191664 GG genotype. At the same time, we further analyzed the linkage disequilibrium and haplotype of three SNP loci in NEIL2 gene. There was no linkage disequilibrium among the three SNPs each other. However, we noted that there may be a trend of linkage disequilibrium between rs8191613 and rs8191664 in CIN III group, while in CSSS group, there may be a trend of linkage disequilibrium between rs804270 and rs8191664.
In summary, these results suggested that two genetic polymorphisms (rs804270 and rs8191664) in the NEIL2 gene were associated with susceptibility to CIN III and CSCC. This effect is likely to be due to alterations in NEIL2 repair activity arising from a change in protein expression or functional domain structure. The GC-GT, CC-GG and CC-GT genotypes at rs804270, and rs8191664 SNPs in the NEIL2 gene, may act as a genetic biomarker to predict the susceptibility to CIN III and CSCC.
Methods
Subject selection and sexual, reproductive, and HR-HPV infection history characteristics
Four hundred CSCCs, four hundred CIN III and one thousand and two hundred normal healthy controls were selected for this study from Chinese population. Their pathological diagnosis was confirmed by two gynecologic pathologists. Normal, healthy female volunteers served as controls and were recruited during gynecological examinations from 2004 to 2008. Normal healthy controls were selected according to the criteria of no pathological cytology findings, endometriosis, gynecological neoplasm, and other solid tumors or immune diseases. Of these, 201 CSCC patients, 357 CIN III patients and 609 normal healthy controls agreed to obtain cervical brushing exfoliated cells to do HR-HPV detection.The infection rates of HR-HPV in CSCC, CIN III and normal healthy controls group were 88.6%, 86.8% and 31.4% respectively. The infection rate of HR-HPV in patients with CIN III and CSCC was significantly higher than that in healthy controls (P < 0.001, χ2 = 277.1; P < 0.001, χ2 = 199.3, respectively).
In normal healthy control group, CIN III group and CSCC group, the number of patients younger than or older than 40 years old was 602/598, 258/142 and 160/240, respectively. Compared with the normal healthy control group, the age of CSCC group was significantly higher than that of 40 years old (P < 0.001, χ2 = 12.4), while the age of CIN III group was lower than that of 40 years old (P < 0.001, χ2 = 24.7). In CIN III and CSCC groups, more individuals with more than three parities were found(P = 0.031, χ2 = 4.6; P < 0.001, χ2 = 20.5, respectively). In CSCC, CIN III and the normal healthy control group, stratified analysis by age at the time of first sexual intercourse (patients were grouped under 20 years old or over), number of sexual partners (patients were grouped by one or more partners) and age at the time of first birth (patients were grouped under 20 years old) showed that there was no statistical difference in this stratification within the group.
Ethical statement
This study was approved by the Medical Ethics Committee of Women’s Hospital Affiliated to Medical School of Zhejiang University (No. 2004002). Informed consent was signed by both patients and normal controls. All the research methods protocols were followed under the approved guidelines and regulations.
SNP selection
We searched for SNPs in the NEIL1 and NEIL2 genes from SNP Library Established by National Library of Medicine (website: www.ncbi.nlm.nih.gov). By utilizing filters (SNP, minor allele frequency (MAF) from 0.05 to 0.5), we obtained seven effective SNPs in the NEIL1 gene. Interestingly, these seven SNPs were located in introns. By utilizing filters for the NEIL2 gene (SNP, missense, MAF from 0.05 to 0.5), we only obtained three effective SNPs in the NEIL2 gene.
The ten SNPs are listed as follows: rs4462560 (C/G), rs7182283 (G/T), rs7402844 (C/G), rs5745920 (C/T), rs8030014 (A/G), rs11634109 (C/T), and rs79244935 (C/T) in the NEIL1 intronic region; rs804270 (C/G) in the NEIL2 5′ UTR region, rs8191613 (A/G) in the NEIL2 intronic region, and rs8191664 (G/T) in the NEIL2 exonic region.
gDNA extraction and SNP genotyping
According to the manufacturer’s protocol, we use the whole genome DNA(gDNA) extraction kit to extract genomic DNA from anticoagulant peripheral blood. (Sangon Bio Co., Shanghai, China). Genomic DNA dissolves in deionized water and is cryopreserved.
Ten SNP genotypes in NEIL1 and NEIL2 genes were determined by modified allele mismatch amplification polymerase chain reaction (MAMA-PCR), as described earlier41. Specific forward and reverse primers and product lengths for MAMA-PCR are shown in Table S1.
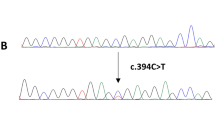
Briefly, the PCR reaction was carried out in a total 20 µL volume reaction mixture containing 20 ng gDNA, 5.0 pmol forward and reverse primer, 0.25 mm dNTP and 1.0U Taq DNA polymerase (TAKARA Co., Dalian, China).The conditions of PCR reaction were as follows: initial denaturation at 94 °C for 5 minutes, followed by 35 cycles: denaturation at 94 °C for 30 seconds, annealing at 55–58 °C for 30 seconds (different primer pairs required different annealing temperatures), and elongation at 72 °C for 30 seconds. At last, a final elongation at 72 °C was performed for 10 minutes. PCR products were analyzed by 2% agarose gel electrophoresis followed by ethidium bromide staining. All the results were measured twice by two technicians with double blind method, and the repeatability of the experiment was completely consistent. In order to further verify the reliability of MAMA-PCR, we selected 5 samples of three genotypes of each locus for using DNA sequencing. In our study, there are 10 loci in total, so the total number of sequencing is: 10 loci * 3 genotypes * 5 samples = 150. The sequencing results of these 150 cases are identical with those of MAMA-PCR. The electropherogram was shown in Fig. S1.
Detection of HR-HPV infection
Hybrid Capture II kit(Digene Diagnostics Co., USA) with probe B was used to detect HR-HPV infection. Probe B can detect 13 subtypes of HR-HPV in total (including 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68). The cervical exfoliated cells for testing were obtained using Digene cervical sampler according to the manufacturer’s instructions.
Detection of NEIL2 mRNA expression
Ninety-two freshly-frozen CSCC tissue samples were used for RNA isolation and NEIL2 gene expression analysis. According to the manufacturer’s procedure, TRIzol reagent(Invitrogen Co., USA) was used to extract total RNA from tissues. The total RNA of each sample was digested by RNase-free DNase I. The purity and quantity of RNA was confirmed with a NanoDrop 2000 (Thermo Fisher). Absorbance at 260/280 of total RNA was between 1.8 and 2.0. The synthesized cDNA serves as a template for qRT-PCR to detect mRNA expression. The reaction conditions of qRT-PCR were 95 °C 30 seconds, followed by 40 cycles: 95 °C, 5 seconds; 60 °C, 35 seconds. The primer sequences for detecting NEIL2 mRNA (NM_001135746.2) were 5′-ATGGAAAGAAATTATTCCTT-3′; and 5′-CAGAATCATCCTCGCCCTGG-3′. GAPDH was served as an internal reference for qRT-PCR. The primer sequences of GAPDH mRNA were 5′-GAGAAGGCTGGGGCTCATTT-3′ and 5′-AGTGATGGCATGGACTGTGG-3′. The length of PCR products of GAPDH and NEIL2 were 231 bp and 204 bp, respectively. All the PCR reactions were performed on ABI’s VIIA 7 DX system. The ΔCt for NEIL2 mRNA expression was calculated compared with the Ct of internal reference GAPDH. The mRNA expression of NEIL2 was calculated by formula: 2−ΔΔCt.
Immunoblotting for NEIL2 protein
NEIL2 protein expression was detected in 92 CSCC tissue samples by immunoblotting. Simply, the tissue sample was minced on ice, dissolved in RIPA tissue lysate buffer, and then homogenized. The supernatant was collected and the protein concentration was detected after rotating the test tube at 4 °C for 1 hour and centrifuging at 12,000 rpm at 4 °C.
Protein lysate (10 μL) was electrophoretic separated on 8% polyacrylamide gel, and then the imprinted proteins were transferred to 0.45 µm PVDF membranes. The PVDF membrane was cultured overnight with the primary antibodies NEIL1 (1:2000) (Proteintech Co., USA) NEIL2 (1:1000) (Invitrogen Co., USA), and GAPDH (1:5000) (Proteintech Co., USA) at 4 °C after 1 hour blocking with 5% nonfat-milk. The membrane was washed three times with TBS buffer containing 0.05% Tween-20, and then incubated for 1 hour with an HRP-conjugated secondary antibody. After fluorescent labeling with ECL substrate, Image Quant LAS 4000 mini (GE Healthcare Co., USA) was used to image the ECL membranes, and then quantitative analysis of the proteins was performed.
Statistical analysis
In order to analyze the correlation between genotype and the risk of CSCC, binary logistic regression analysis was used to obtain odds ratio (OR), 95% confidence interval (CI) and P value. The normal control group was acted as a reference. FDR adjusted p values were corrected by the method of Benjamin Hochberg (BH method) for multiple testing correction. Kruskal-Wallis H test was used for stratified analysis of reproductive and sexual history and genotype distribution frequency. Multinomial regression analysis was performed among the different groups for different genotypes, and less than 0.05 of P value indicates the model fitting is significant. To analyze the differences in the expression of mRNA or protein, ANOVA (Fisher’s Least Difference test) or Student’s two-tailed t-test was used for statistical analysis. Statistical significance level was set at P ≤ 0.05, and it was a bilateral test. All statistical processes are completed by SPSS software (Version 18.0 for Windows). The linkage disequilibrium of three SNP loci in NEIL2 gene was analyzed with the SHEsis software42.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 (2016).
Walboomers, J. M. et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19 (1999).
Arbyn, M. et al. Worldwide burden of cervical cancer in 2008. Ann. Oncol. 22, 2675–86 (2011).
Chen, W. et al. Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115–132 (2016).
Bosch, F. X. et al. Prevalence of human papillomavirus in cervical cancer: a worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J. Natl Cancer Inst. 87, 796–802 (1995).
Zhang, X., Zhang, L., Tian, C., Yang, L. & Wang, Z. Genetic variants and risk of cervical cancer: epidemiological evidence, meta-analysis and research review. BJOG 121, 664–674 (2014).
Chen, D. et al. Genome-wide association study of susceptibility loci for cervical cancer. J. Natl Cancer Inst. 105, 624–633 (2013).
Shi, Y. et al. A genome-wide association study identifies two new cervical cancer susceptibility loci at 4q12 and 17q12. Nat. Genet. 45, 918–922 (2013).
Sancar, A. DNA excision repair. Annu. Rev. Biochem. 65, 43–81 (1996).
Hazra, T. K. et al. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J. Biol. Chem. 277, 30417–30420 (2002).
Hazra, T. K. & Mitra, S. Purification and characterization of NEIL1 and NEIL2, members of a distinct family of mammalian DNA glycosylases for repair of oxidized bases. Methods Enzymol. 408, 33–48 (2006).
Bandaru, V., Sunkara, S., Wallace, S. S. & Bond, J. P. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 1, 517–529 (2002).
Das, A. et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair. 5, 1439–1448 (2006).
Zhai, X. et al. Functional variants of the NEIL1 and NEIL2 genes and risk and progression of squamous cell carcinoma of the oral cavity and oropharynx. Clin. Cancer Res. 14, 4345–4352 (2008).
Goto, M., Shinmura, K., Tao, H., Tsugane, S. & Sugimura, H. Three novel NEIL1 promoter polymorphisms in gastric cancer patients. World J. Gastrointest. Oncol. 2, 117–120 (2010).
Wei, H. et al. Association of polymorphisms in oxidative stress genes with clinical outcomes for bladder cancer treated with Bacillus Calmette-Guérin. PLoS One 7, e38533 (2012).
Corral, R. et al. Genetic variation in the base excision repair pathway, environmental risk factors, and colorectal adenoma risk. PLoS One 8, e71211 (2013).
Götz, M. E., Künig, G., Riederer, P. & Youdim, M. B. Oxidative stress: free radical production in neural degeneration. Pharmacol. Ther. 63, 37–122 (1994).
Grisham, M. B., Granger, D. N. & Lefer, D. J. Modulation of leukocyte-endothelial interactions by reactive metabolites of oxygen and nitrogen: relevance to ischemic heart disease. Free. Radic. Biol. Med. 25, 404–433 (1998).
Breen, A. P. & Murphy, J. A. Reactions of oxyl radicals with DNA. Free. Radic. Biol. Med. 18, 1033–1077 (1995).
Lord, C. J. & Ashworth, A. The DNA damage response and cancer therapy. Nat. 481, 287–294 (2012).
Halazonetis, T. D., Gorgoulis, V. G. & Bartek, J. An oncogene-induced DNA damage model for cancer development. Sci. 319, 1352–1355 (2008).
Bonn, D. How DNA-repair pathways may affect cancer risk. Lancet 351, 42 (1998).
Wood, R. D., Mitchell, M., Sgouros, J. & Lindahl, T. Human DNA repair genes. Sci. 291, 1284–1289 (2001).
Dizdaroglu, M., Coskun, E. & Jaruga, P. Repair of oxidatively induced DNA damage by DNA glycosylases: Mechanisms of action, substrate specificities and excision kinetics. Mutat. Res. 771, 99–127 (2017).
Dianov, G. L. & Hübscher, U. Mammalian base excision repair: the forgotten archangel. Nucleic Acids Res. 41, 3483–3490 (2013).
Lindahl, T. Instability and decay of the primary structure of DNA. Nat. 362, 709–715 (1993).
Krokan, H. E., Standal, R. & Slupphaug, G. DNA glycosylases in the base excision repair of DNA. Biochem. J. 325, 1–16 (1997).
Hazra, T. K. et al. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA 99, 3523–3528 (2002).
Rosenquist, T. A. et al. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair. 2, 581–591 (2003).
Takao, M. et al. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J. Biol. Chem. 277, 42205–42213 (2002).
Dou, H., Mitra, S. & Hazra, T. K. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 278, 49679–49684 (2003).
Vartanian, V. et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl Acad. Sci. USA 103, 1864–1869 (2006).
Hazra, T. K. et al. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 6, 470–480 (2007).
Shinmura, K. et al. Abnormal Expressions of DNA Glycosylase Genes NEIL1, NEIL2, and NEIL3 Are Associated with Somatic Mutation Loads in Human Cancer. Oxid. Med. Cell Longev. 2016, 1546392 (2016).
Chae, Y. K. et al. Genomic landscape of DNA repair genes in cancer. Oncotarget 7, 23312–23321 (2016).
Elingarami, S. et al. Polymorphisms in NEIL-2, APE-1, CYP2E1 and MDM2 Genes are Independent Predictors of Gastric Cancer Risk in a Northern Jiangsu Population (China). J. Nanosci. Nanotechnol. 15, 4815–4828 (2015).
Dey, S. et al. Increased risk of lung cancer associated with a functionally impaired polymorphic variant of the human DNA glycosylase NEIL2. DNA Repair. 11, 570–578 (2012).
Sanders, A. R. et al. Genome-Wide Association Study of Male Sexual Orientation. Sci. Rep. 7, 16950 (2017).
Pearce., E., Wlodarski, R., Machin, A. & Dunbar, R. I. M. The Influence of Genetic Variation on Social Disposition, Romantic Relationships and Social Networks: a Replication Study. Adapt. Hum. Behav. Physiol. 4, 400–422 (2018).
Ye, F., Cheng, Q., Hu, Y., Zhang, J. & Chen, H. PARP-1 Val762Ala polymorphism is associated with risk of cervical carcinoma. PLoS One 7, e37446 (2012).
Shi., Y. Y. & He., L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 15, 97–8 (2005).
Acknowledgements
We would like to thank Dr. Zhou Caiyun and Dr. Yu Minhua for their help in recruiting subjects. In addition, we would like to thank the International Science Editor (http://www.international science editing.com) for the language editing of our manuscript. This project was supported by grants from Zhejiang Provincial Natural Science Foundation of China (No. Y2110200) and the National Nature Science foundation of China (No. 30973380).
Author information
Authors and Affiliations
Contributions
Conceived and designed this project: H.C. F.Y. Finished the experiments: F.Y., H.W., Q.C., X.C., J.L. Data statistics: F.Y., H.C. Drafting, revising and finalizing the paper: F.Y., H.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ye, F., Liu, J., Wang, H. et al. Cervical carcinoma risk associate with genetic polymorphisms of NEIL2 gene in Chinese population and its significance as predictive biomarker. Sci Rep 10, 5136 (2020). https://doi.org/10.1038/s41598-020-62040-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62040-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.