Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive malignancies. TGF-β is strongly expressed in both the epithelial and stromal compartments of PDAC, and dysregulation of TGF-β signalling is a frequent molecular disturbance in PDAC progression and metastasis. In this study, we investigated whether blockade of TGF-β signalling synergizes with nal-IRI/5-FU/LV, a chemotherapy regimen for malignant pancreatic cancer, in an orthotopic pancreatic tumour mouse model. Compared to nal-IRI/5-FU/LV treatment, combining nal-IRI/5-FU/LV with vactosertib, a TGF-β signalling inhibitor, significantly improved long-term survival rates and effectively suppressed invasion to surrounding tissues. Through RNA-sequencing analysis, we identified that the combination treatment results in robust abrogation of tumour-promoting gene signatures and positive enrichment of tumour-suppressing and apoptotic gene signatures. Particularly, the expression of tumour-suppressing gene Ccdc80 was induced by vactosertib and further induced by vactosertib in combination with nal-IRI/5-FU/LV. Ectopic expression of CCDC80 suppressed migration and colony formation concomitant with decreased expression of epithelial-to-mesenchymal transition (EMT) markers in pancreatic cancer cells. Collectively, these results indicate that combination treatment of vactosertib with nal-IRI/5-FU/LV improves overall survival rates in a mouse model of pancreatic cancer by suppressing invasion through CCDC80. Therefore, combination therapy of nal-IRI/5-FU/LV with vactosertib could provide clinical benefits to pancreatic cancer patients.
Similar content being viewed by others
Introduction
Pancreatic cancer is one of the leading causes of cancer-related mortality in the world. The 5-year relative survival rate of pancreatic ductal adenocarcinoma (PDAC) patients was only 8% for all stages combined data in 20181. Although 5-year relative survival rates in other cancer types have steadily increased over the years, improvement in pancreatic cancer is very slow because patients with pancreatic cancer are often diagnosed at advanced stages due to absence of noticeable symptoms in the early stages2,3. Moreover, control of pancreatic cancer is difficult owing to its aggressiveness, distal metastasis, resistance to most common treatments, and genetic and epigenetic alterations4,5,6.
The pancreatic cancer treatment paradigm has improved in the last 20 years. In clinical trials in the 1970s, 5-fluorouracil (5-FU) was used after resection7. Nowadays, the combination treatments such as FOLFIRINOX (oxaliplatin, irinotecan, fluorouracil (5-FU), and leucovorin (LV)) and gemcitabine plus nab-paclitaxel are the novel chemotherapy regimens used for patients with metastatic pancreatic adenocarcinoma (mPAC), displaying significantly better patient outcomes compared to the commonly used gemcitabine and providing the chance for salvage chemotherapy8,9,10. Nanoliposomal irinotecan (nal-IRI) has different pharmacokinetic properties from irinotecan owing to the outer PEGylated liposomes encapsulating the irinotecan sucrosofate salt. This very property of nal-IRI allows increased and prolonged levels of irinotecan and its metabolite SN-38 to carry out anti-tumour activity with low toxicity compared to free-irinotecan11. Based on the phase 2 clinical study of nal-IRI against pancreatic cancer12, the pivotal phase III NAPOLI-1 study has been conducted in gemcitabine-failed pancreatic cancer patients. In the NAPOLI-1 study, the combination of nal-IRI and 5-FU/LV significantly increased the median survival (6.1 months) compared to the 5-FU/LV control (4.2 months) or nal-IRI monotherapy (4.9 months)13. In this clinical trial, the effectiveness of nal-IRI is improved by co-treatment with 5-FU/LV rather than monotherapy of each regimen.
Regulation of TGF-β signalling is essential for controlling tumour progression and metastasis because TGF-β is involved in cell proliferation, epithelial-to-mesenchymal transition (EMT), angiogenesis, metastasis, and immune responses in cancer14,15. For example, when the TGF-βRI/II kinase inhibitor LY-2109761 was co-administered with gemcitabine in an orthotopic xenograft mouse model of pancreatic cancer, tumour growth and spontaneous metastasis were significantly reduced with improved median survival16. Loss of TGF-β signalling induced by SB-431542, an inhibitor of TGF-β type I receptor, increased sensitivity to rapamycin-induced apoptosis in MDA-MB-231-derived breast cancer17. Vactosertib (TEW-7197) is an orally bioavailable TGF-β signalling inhibitor that also targets the TGF-β type I receptor kinase18,19. Treatment of vactosertib reduces cancer cell migration, invasion, and metastasis of diverse cancers, including breast cancer, pancreatic cancer, and melanoma20,21,22. Recently, clinical studies of vactosertib-combined treatment with other anti-cancer drugs are ongoing in various cancer types. Particularly, vactosertib and FOLFOX combination therapy is undergoing evaluation for mPAC patients. Therefore, vactosertib could be an attractive candidate for pancreatic cancer therapy considering its ability to inhibit TGF-β signalling in vivo.
In this study, we investigated whether vactosertib manifests therapeutic benefits to pancreatic cancer patients by evaluating the effects of combination treatment of vactosertib with nal-IRI/5-FU/LV in an orthotopic mouse model of pancreatic cancer. We found that the combination dramatically increased the survival rate by suppressing invasion of pancreatic cancer cells. Through RNA-sequencing analysis, we found that genes involved in the regulation of the apoptotic process might be pertinent to the improved survival rate. Among these genes, we selected CCDC80, a tumour suppressor gene that was significantly induced by the combination treatment, to further investigate the underlying mechanism of the combination treatment. Indeed, ectopic expression of CCDC80 suppressed migration, colony formation, and expression of EMT markers. In summary, the combination of vactosertib with nal-IRI/5-FU/LV improved survival in the pancreatic cancer mouse model by inhibiting migration, invasion, and EMT of pancreatic cancer cells.
Results
Combination of vactosertib with nal-IRI/5-FU/LV significantly improved survival in a mouse pancreatic cancer model
Prior to the investigation of the effects of combination of vactosertib with nal-IRI/5-FU/LV, we first examined whether blockade of TGF-β signalling through vactosertib, targeting TGF-β type I receptor, is beneficial for the treatment of pancreatic cancer by analysing the expression of TGFBR1 in normal (n = 171) and pancreatic tumour (n = 179) tissues using TCGA database. We found that TGFBR1 was significantly overexpressed in tumour tissues (Supplementary Fig. 1A). We then tested the effect of vactosertib on PANC-1 human pancreatic cancer cell. As shown in Supplementary Fig. 1B,C, vactosertib treatment restored TGF-β1-mediated suppression of CDH1 and reduced TGF-β1-mediated increase of CDH2, SNAI1, and SNAI2, ultimately suppressing TGF-β1-induced migration of PANC-1 cells. Based on these observations, we investigated whether vactosertib co-administration enhances the anti-tumour effects of nal-IRI/5-FU/LV in an orthotopic pancreatic cancer mouse model using murine Panc02 pancreatic cancer cells (Fig. 1A). We observed that both nal-IRI/5-FU/LV and Vac + nal-IRI/5-FU/LV effectively suppressed tumour growth (Supplementary Fig. 2A). Both vactosertib and nal-IRI/5-FU/LV monotherapy enhanced survival, but the survival rate was significantly improved by more than 60% in the Vac + nal-IRI/5-FU/LV-administered group (Fig. 1B). Interestingly, as demonstrated by H&E staining, borders between tumour tissues and pancreatic parenchyma were clearly demarcated in the vactosertib alone- and Vac + nal-IRI/5-FU/LV-administered groups (Fig. 1C). Indeed, cancer cells in the vactosertib- and Vac + nal-IRI/5-FU/LV-administered groups showed epithelial phenotype, while cells in the control and nal-IRI/5-FU/LV-administered groups exhibited mesenchymal phenotype with a spindle-like shape (Fig. 1D). Immunohistochemical analysis also demonstrated that the expression of vimentin, a representative EMT marker, was markedly reduced in both vactosertib alone- and Vac + nal-IRI/5-FU/LV-administered groups (Fig. 1E). These results suggest that both vactosertib and its combination with nal-IRI/5-FU/LV might reduce invasiveness and EMT of pancreatic cancer. Furthermore, we observed that administration of nal-IRI/5-FU/LV alone could induce fibrotic changes such as accumulation of α-smooth muscle actin (α-SMA) and deposition of collagen in the pancreatic tumour tissues that might interfere with the function of pancreatic parenchyma adjacent to cancer (Supplementary Fig. 2B). However, combination treatment of vactosertib with nal-IRI/5-FU/LV significantly suppressed fibrotic changes in the pancreatic tumour tissues by reducing α-SMA expression and collagen deposition. Taken together, combination treatment of vactosertib with nal-RI/5-FU/LV demonstrated favourable therapeutic effects on the syngeneic mouse model of pancreatic cancer through reduction of primary tumour sizes and suppression of invasion to surrounding tissues.
Survival improvement in response to combination treatment of vactosertib with nal-IRI/5-FU/LV in the orthotopic pancreatic cancer mouse model. (A) Experimental design of C57BL/6 syngeneic orthotopic mouse model using Panc02 murine pancreatic cancer cells. Five days after cell injection (3 × 106), mice were randomized into 4 groups. Vactosertib was administered orally for 5 consecutive days followed by 2 days of resting period starting from day 6 post cell injection. nal-IRI/5-FU/LV was injected intraperitoneally every 2 weeks starting from day 8 post cell injection. (B) Relative survival rates of the control, vactosertib, nal-IRI/5-FU/LV, and combined treatment groups (***P < 0.0005 and **P < 0.005 compared to the control group) (C) Haematoxylin and eosin (H&E) staining of pancreatic tumour tissues showing tumour cell invasion to adjacent pancreas tissues. Black dotted lines indicate the borders between tumourous and normal pancreas without invading tumour cells. (D) The morphologies of tumour cells presented by H&E staining. Yellow arrows point out the representative mesenchymal (control and nal-IRI/5-FU/LV) and epithelial (vactosertib and Vac + nal-IRI/5-FU/LV) cell morphologies in the images. (E) The immunohistochemical staining of vimentin in tumour tissues of each group.
In addition, we examined whether combination of vactosertib with nal-IRI monotherapy increases survival rates, even though nal-IRI monotherapy showed lower efficiency than nal-RI/5-FU/LV in the clinical trial13. Compared to either nal-IRI or vactosertib monotherapy, combination of vactosertib with nal-IRI resulted in a significantly prolonged survival rate in the mouse model of pancreatic cancer (Supplementary Fig. 3A,B). Similar to Vac + nal-IRI/5-FU/LV-administered mice, histological analysis showed increased epithelial phenotype and reduced tumour cell invasion in vactosertib-administered groups (Supplementary Fig. 3C), suggesting that the combination of vactosertib with nal-IRI also improves survival rates and reduces invasiveness of pancreatic cancer in the mouse model.
Co-treatment of vactosertib with nal-IRI/5-FU suppresses migration, invasion, and EMT of pancreatic cancer cells
Based on in vivo observations, we next examined the combined effect of vactosertib with nal-IRI/5-FU in pancreatic cell lines. Because leucovorin (LV) is used as an adjuvant chemical to reduce the toxicity of anti-cancer drugs in vivo, we only used 5-FU for in vitro experiments. In addition to Panc02 cells that were used for in vivo experiments, we selected PANC-1 and SNU2491 human pancreatic cancer cell lines that showed responses to TGF-β1 for further investigation (Supplementary Fig. 4). In order to examine the effect of vactosertib on invasiveness of pancreatic cancer cells in vitro, we first performed migration and invasion assays. Each vactosertib and nal-IRI/5-FU treatment significantly reduced migration of PANC-1, Panc02, and SNU2491 cells while the combination treatment of vactosertib with nal-IRI/5-FU reduced migration even further (Fig. 2A and Supplementary Fig. 5A). The synergistic effect of vactosertib and nal-IRI/5-FU was further investigated with invasion assay using a chamber coated with matrigel that mimics extracellular matrix. Compared to the pancreatic cancer cells of the control, vactosertib, and nal-IRI/5-FU groups that had extendedly degraded matrigel and migrated through the chamber, the combination treatment reduced invasive capability of pancreatic cancer cells (Fig. 2B and Supplementary Fig. 5B). We also studied the effect of vactosertib and its combination with nal-IRI/5-FU by examining EMT responses. First of all, PANC-1, Panc02, and SNU2491 cells all showed TGF-β1-induced EMT showing reduced expression of epithelial marker E-cadherin as well as increased expression of mesenchymal markers such as N-cadherin, Fibronectin, and Snail. Compared to nal-IRI/5-FU, vactosertib effectively reduced the TGF-β1-mediated induction of mesenchymal marker expression and restored the TGF-β1-mediated reduction of E-cadherin expression. However, combination treatment of vactosertib with nal-IRI/5-FU significantly ameliorated TGF-β1-induced EMT of these pancreatic cancer cells (Fig. 2C and Supplementary Fig. 5C). These data suggest that when combined with nal-IRI/5-FU, vactosertib exerts synergistic effects on suppressing migration and invasion of pancreatic cancer cell lines by regulating EMT.
Synergistic effect of vactosertib with nal-IRI/5-FU on migration, invasion, and EMT of pancreatic cancer cells. (A) Transwell cell migration assay. Cells were treated with the indicated reagent(s) and incubated for 48 hours before placed in a migration chamber. 1 × 105 of PANC-1 (left) or 5 × 104 of Panc02 (right) cells were then seeded in migration chambers for 48 hours. The relative number of migrated cells in each group was counted. (B) Cell invasion assay. Cells were treated with the indicated reagent(s) and incubated for 48 hours before placed in an invasion chamber. 2 × 105 PANC-1 (left) or 1 × 105 Panc02 (right) cells were then seeded in invasion chambers for 48 hours. The number of invaded cells in each group was counted. Note that the combination of vactosertib with nal-IRI/5-FU inhibits migration and invasion in PANC-1 and Panc02 pancreatic cancer cells. (A,B) The values for migrated/invaded number of cells represent the mean ± SD of triplicate data. ***P < 0.0005, **P < 0.005, and *P < 0.05 compared to the control group; ##P < 0.005 and #P < 0.05 compared to the nal-IRI-treated group. (C) Western blot analysis for EMT markers. Vactosertib was treated for 2 hours prior to incubation with nal-IRI/5-FU in the presence of 5 ng/ml TGF-β for 48 hours. The blots are cropped and the full-length images are presented in Supplementary Fig. 8. Note that vactosertib and its combination with nal-IRI/5-FU restore the TGF-β-mediated reduction of E-cadherin and the increase of mesenchymal markers.
Transcriptome profiling of pancreatic tumour tissues following combination treatment of vactosertib with nal-IRI/5-FU/LV
To further explore the mechanism by which combination treatment of vactosertib with nal-IRI/5-FU/LV has on the survival of the pancreatic tumour mouse model, we performed RNA-seq analysis of the pancreatic tumours. The sequencing analysis yielded an average of 89.7 (±25.9) million reads per library (Additional file 1). Compared to the control tumours, we identified 571, 648, and 976 differentially expressed genes (DEGs) in the pancreatic tumours that had been administered with vactosertib, nal-IRI/5-FU/LV, and the combination treatment, respectively. Among the DEGs, we identified a total of 325 genes (cluster II, III, and IV) that were differentially expressed in response to either vactosertib or nal-IRI/5-FU/LV monotherapy (Fig. 3A). Interestingly, these genes were significantly regulated by the combination treatment and exhibited a reverse expression pattern between the control and the Vac + nal-IRI/5-FU/LV-administered groups. To gain a better understanding of the involvement of these genes in the effects of combination treatment in pancreatic cancer, functional categories of the genes were subjected to gene ontology (GO) enrichment analysis. Our analysis of the genes revealed significant functions associated with regulation of phosphate metabolic processes (P = 3 × 10−9), cell migration (P = 1.3 × 10−7), positive regulation of apoptotic processes (P = 1.8 × 10−7), vasculature development (P = 3 × 10−7), and positive regulation of transport (P = 1.1 × 10−7) (Fig. 3B, Supplementary Table 1). In particular, enrichment of the genes such as Sfrp1, Sfrp2, Il1b, Lpar1, Pmaip1, Ctla4, Cyp1b1, Map3k5, Nfatc4, Aldh1a2, Nr4a1, Clip3, and Dusp1 in the functional category of positive regulation of apoptotic process was supported by gene set enrichment analysis (GSEA) (Fig. 3C). These findings suggest that the combination treatment might exert an effect on the apoptotic process of pancreatic cancer. To confirm whether the combination treatment regulates apoptosis, we performed TUNEL assay using tumour tissue sections. As shown in Fig. 3D, the apoptotic cells were significantly increased in Vac + nal-IRI/5-FU/LV-administered tumour tissues. Taken together, our results indicate that combination treatment of vactosertib with nal-IRI/5-FU/LV might reduce pancreatic tumour progression not only by suppressing invasion, but also by enhancing apoptosis while simultaneously regulating DEGs.
RNA-sequencing analysis of pancreatic tumour tissues obtained from orthotopic mouse model administered with vactosertib, nal-IRI/5-FU/LV, and the combination. (A) Heatmap plot of DEGs using the hierarchical clustering method. (B) Top 10 pathways characterized by from GO enrichment analysis of DEGs (cutoff of P < 0.001). (C) GO enrichment plot for positive regulation of apoptotic process plotted by Gene Set Enrichment Analysis (GSEA). (D) TUNEL assay of pancreatic tumour tissues used for RNA sequencing. Note that more apoptotic cells were shown in the tumour tissues obtained from mice administered with combination of vactosertib with nal-IRI/5-FU/LV.
Identification of Ccdc80 and tumorigenesis-related genes regulated by combination treatment of vactosertib with nal-IRI/5-FU/LV
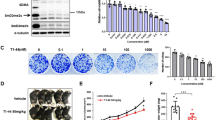
From RNA-Seq analysis, we found that tumour suppressors such as Ccdc80, Itm2a, and Sfrp1 were significantly up-regulated by the combination treatment of vactosertib with nal-IRI/5-FU/LV compared to the control group and the nal-IRI/5-FU/LV group (Fig. 4A and Supplementary Fig. 6A). We further confirmed altered expression levels of the selected genes by quantitative RT-PCR (Fig. 4B and Supplementary Fig. 6B). We found that the expression levels of several oncogenes, including Axl, Tkt, Sncg, and Bcl9l, were down-regulated by the treatment of vactosertib with nal-IRI/5-FU/LV (Supplementary Fig. 6A,B). Consistently, the expression levels of the tumour suppressor genes were higher in the healthy mouse pancreatic tissues than in the pancreatic tumour tissues obtained from the orthotopic mouse model using Panc02 cells. On the other hand, the expression levels of the oncogenes were higher in the tumour tissues than in the healthy tissues (Supplementary Fig. 6C). Among the genes identified, Ccdc80 is one of the known tumour suppressors regulating the expression of E-cadherin23. According to previous studies, CCDC80 is related to cancer cell motility in thyroid, colorectal, and pancreatic cancers24,25. Down-regulation of Ccdc80 is detected in pancreatic cancer cell lines and primary tumours, and ectopic expression of CCDC80 promotes cell growth inhibition and sensitizes cells to apoptosis in colon and pancreatic cancer cell lines25. Interestingly, Ccdc80 was induced by vactosertib, and it was further induced by the combination treatment of vactosertib with nal-IRI/5-FU/LV. Through immunohistochemical staining of Ccdc80, we confirmed that the protein expression of Ccdc80 was significantly increased in the cancer tissues obtained from the vactosertib and Vac + nal-IRI/5-FU/LV-administered mice compared to the ones from the control and nal-IRI/5-FU/LV-administered mice (Fig. 4C). We also analysed mRNA levels of CCDC80 in the vactosertib and nal-IRI/5-FU-treated PANC-1 and Panc02 cell lines. Since the expression of CCDC80 was suppressed by TGF-β1, inhibition of TGF-β signalling by vactosertib significantly induced CCDC80 expression, which was further induced by vactosertib in combination with nal-IRI/5-FU (Fig. 4D). These results suggest that CCDC80 might be an important mediator for combination treatment of vactosertib with nal-IRI/5-FU/LV in reducing pancreatic tumour progression.
Up-regulation of CCDC80 expression by vactosertib and its combination with nal-IRI/5-FU/LV. (A) FPKM value of Ccdc80 in the tumour tissues from RNA sequencing. (B) qRT-PCR result validating Ccdc80 mRNA expression in mouse tumour tissues. The values represent the mean ± SD of triplicate samples. **P < 0.005 compared to the control group. ###P < 0.0005 compared to the nal-IRI/5-FU/LV group. (C) Immunohistochemical staining of mouse tumour tissues with CCDC80. Note that CCDC80 expression is increased in the tumour tissues from the mice administered with vactosertib or its combination with nal-IRI/5-FU/LV. (D) RT-PCR analysis revealing up-regulation of CCDC80 by vactosertib and its combination with nal-IRI/5-FU in PANC-1 and Panc02 cell lines.
Ectopic expression of CCDC80 in pancreatic cancer cells decreases migration, colony formation, and EMT
We further investigated whether CCDC80 plays a role in EMT of pancreatic cancer cells. Consistent with down-regulation of Ccdc80 in the pancreatic tumour tissues, mRNA expression of CCDC80 was significantly decreased in most of the pancreatic cancer cell lines compared to HPDE normal pancreatic cell line (Supplementary Fig. 6C,D). To investigate the role of CCDC80 in pancreatic cancer progression, we generated PANC-1 and Panc02 cells stably expressing 3Flag-CCDC80. We found that the ectopic expression of CCDC80 significantly reduced cell migration and colony formation concomitant with down-regulation of mesenchymal markers such as CDH2, FN, VIM, SNAI1, SNAI2, and ZEB1, and up-regulation of epithelial marker CDH1 in both PANC-1 and Panc02 cells (Fig. 5A–C). Western blot analysis demonstrated that the expression level of E-cadherin was increased and the expression levels of mesenchymal markers such as N-cadherin, Fibronectin, Vimentin, Snail, and Slug were significantly decreased by ectopic expression of CCDC80 (Fig. 5D). The same results were also observed in SNU2491 cells stably expressing CCDC80 (Supplementary Fig. 7). Collectively, these findings suggest that CCDC80 might participate in the underlying mechanism of combination treatment of vactosertib with nal-IRI/5-FU/LV in suppressing invasiveness and subsequent pancreatic cancer progression.
Ectopic expression of CCDC80 reducing migration, colony formation, and EMT in pancreatic cancer cells. (A) Transwell migration assay measuring migration abilities of PANC-1 and Panc02 cells expressing LPCX or CCDC80. (B) Representative images of colonies stained with methylene blue in PANC-1 and Panc02 cells stably expressing CCDC80. (C) qRT-PCR results showing down-regulation of EMT marker expression in PANC-1 and Panc02 cells by ectopic expression of CCDC80. All the data is represented as the mean of three repeated values. ***P < 0.0005, **P < 0.005, and *P < 0.05 compared to the control. (D) Western blot analysis showing reduction of EMT markers in PANC-1 and Panc02 with stably expressing CCDC80. The blots are cropped, and the original blots are presented in Supplementary Fig. 8. Note that overexpression of CCDC80 decreases EMT marker expression in pancreatic cancer cells.
Discussion
TGF-β signalling plays a vital role in pancreatic cancer initiation and progression26. At least one mutation in the TGF-β downstream genes is detected in most pancreatic cancer patients27. Inactivation of a TGF-β signalling mediator leads to a loss of tumour suppressive role of TGF-β in pancreatic cancer28,29. TGF-β plays a dual role during tumorigenesis. It suppresses tumour progression by promoting cell cycle arrest and apoptosis in early-stage of pancreatic carcinogenesis. On the other hand, it functions as a tumour promoter by activating the Smad4-independent signalling pathway, promoting cell motility, invasion, EMT, and metastasis and decreasing anti-tumour immune responses in advanced stages30. Thus, inhibition of TGF-β signalling in the late stages of cancer progression could be an effective strategy for treating pancreatic cancer. In this study, we found that treatment of vactosertib, a TGF-β signalling inhibitor, with either nal-IRI/5-FU/LV or nal-IRI significantly improved overall survival in a mouse model of pancreatic cancer by suppressing EMT and invasion of pancreatic cancer cells. In addition, tumour microenvironment, such as cancer-associated fibroblasts, plays a crucial role in metastasis31. We observed that treatment of vactosertib and its combination with nal-IRI/5-FU/LV effectively reduced α-SMA and collagen deposition in the pancreatic tumour tissues, whereas nal-IRI/5FU/LV treatment induced fibrotic changes that might lead to pancreas dysfunction. Considering the involvement of TGF-β in regulating tumour microenvironment, inhibition of TGF-β through vactosertib might suppress fibrotic changes during tumourigenesis. Taken together, our results suggest that combination treatment of vactosertib with other anti-cancer drugs might be useful in treating advanced-stage pancreatic cancer.
Through RNA-seq analysis, the DEGs related to TGF-β signalling were identified in response to combination treatment of vactosertib with nal-IRI/5-FU/LV. For example, BCL9L and SNCG are involved in TGF-β-induced EMT, migration, invasion, and metastasis32,33. The expression of BCL9L is up-regulated in pancreatic cancer34, and knockdown of BCL9L diminishes TGF-β-induced EMT responses in pancreatic cancer cells and inhibits liver metastasis in vivo32. BCL9L expression level determines the aggressiveness of pancreatic tumours, presenting lower expression in SPN, a benign pancreatic tumour, than in PDAC, an aggressive type34. The expression of SNCG is up-regulated in breast, colon, and pancreatic cancers35,36,37. SNCG is involved in perineural invasion associated with local recurrence and dismal prognosis in relation with TGF-β or Twist in pancreatic cancer33. In this study, we found that combination treatment of vactosertib with nal-IRI/5-FU/LV significantly reduced SNCG and BCL9L expression. Moreover, combination treatment of vactosertib with nal-IRI/5-FU/LV induced well-known tumour suppressors ITM2A and SFRP1. In ovarian cancer, expression level of ITM2A is reduced, and the disease-free survival rate is prolonged in patients in the ITM2A-high patient group38. Loss of SFRP1 has been detected in hepatocellular carcinoma, colon cancer, breast cancer, and many other cancers associated with poor prognosis39,40,41,42. Even though we focused on CCDC80 in the present study, it would be worth investigating whether ITM2A and SFRP1 are involved in the action of combination treatment of vactosertib with nal-IRI/5-FU/LV in ameliorating pancreatic cancer.
According to previous studies, CCDC80 (Dro1) is a secreted protein induced by oestrogen43. It plays a role in osteoblast and adipocyte differentiation, embryonic skeletal development, and glucose homeostasis44,45. In regard to cancer, mRNA expression of CCDC80 is often down-regulated in various cancer cell lines and tumour tissues, suggesting the tumour-suppressive role of CCDC8024,43,46,47. Overexpression of CCDC80 decreases clonogenic capacities of cancer cells, including PANC-1 and BxPC-3 pancreatic cancer cells, and increases CD95-induced apoptosis25. CCDC80 is also known to mediate signals through the FAK/E-cadherin axis, which regulates cell migration23. In addition to these roles of CCDC80, we revealed that the ectopic expression of CCDC80 could also suppress cell migration, colony formation, and EMT in PANC-1, Panc02, and SNU2491 pancreatic cancer cells.
In conclusion, this study demonstrates that combination treatment of vactosertib with nal-IRI/5-FU/LV improves pancreatic cancer survival by suppressing cell migration, invasion, and EMT, highlighting a potential clinical application of this combination therapy for PDAC patients.
Materials and Methods
Cell lines
The human pancreatic cancer cell line PANC-1 was obtained from American Type Culture Collection (ATCC, VA, USA), and the C57BL/6 syngeneic pancreatic adenocarcinoma cell line Panc02 was provided from Medpacto (Korea). Both cell lines were maintained in DMEM with high glucose (WelGENE), supplemented with 10% FBS (WelGENE), 100 U/ml penicillin and 100 μg/ml streptomycin.
Reagents
Vactosertib was obtained from Medpacto (Korea) and solubilized in gastric fluid. Nanoliposomal irinotecan (Onivyde) was a kind gift from Prof. Joon Oh Park from Samsung Seoul Hospital, and 5-fluorouracil was purchased from MCE (Cat. No. HY-90006/CS-0993) and leucovorin (calcium folinate) from TCI (Cat. No. C2235). Both 5-FU and LV were solubilized in PBS. Antibodies against E-cadherin (Cat. No. 610181), N-cadherin (Cat. No. 610921), and Fibronectin (Cat. No. 610077) were obtained from BD Biosciences. Antibodies against Vimentin (Abcam, Cat. No. ab9978), Snail (Cell Signaling, Cat. No. 3879), Slug (Santa Cruz, Cat. No. sc-15391), and CCDC80 (Bioss, Cat. No. bs-7992R) were diluted in 3% BSA solution. Anti-mouse and anti-rabbit secondary antibodies were purchased from Millipore.
TCGA analysis
TCGA analysis for TGFBR1 expression data was obtained from the GEPIA database in human normal pancreatic tissues (N = 171) and pancreatic cancer tissues (N = 179) (http://gepia.cancer-pku.cn). The P value was calculated by unpaired two-tailed Student’s t-tests.
Syngeneic murine model of pancreas orthotopic tumour injection
Six-week-old male immunocompetent C57BL/6 mice were purchased from Japan SLC, Inc. We injected 3 × 106 Panc02 cells with a 1:1 ratio of Matrigel (Corning, Cat. No. 356231) into the tail of pancreas. Five days after implantation, mice were randomized into four treatment groups: control, vactosertib, nal-IRI/5-FU/LV, and combination of vactosertib and nal-IRI/5-FU/LV (n = 10–15 per group). The mice were treated with 25 mg/kg vactosertib in gastric fluid by oral gavage five times a week and/or 10 mg/kg nal-IRI, 25 mg/kg 5-FU, and 45 mg/kg leucovorin via intraperitoneal (i.p.) injection once in two weeks. The control group was injected with gastric fluid (orally) and PBS (i.p.) at the same time. We followed up on the body weight to detect the health of mice and tracked the death of mice for survival. We collected tumour samples from each group at the same time. All animal experiments were approved by the Woojung Bio Animal facility (Suwon, Korea) and all methods were performed in accordance with the relevant guidelines and regulations.
Tumour tissue fixation and immunohistochemistry
Pancreatic tumour tissues were fixed in 10% neutral buffered formalin overnight and then embedded in paraffin blocks. Slides were stained with haematoxylin and eosin (H&E) and Masson’s trichrome. The antibodies used for immunohistochemistry (IHC) were α-SMA (Novus, Cat. No. NBP1-30894), vimentin (Abcam, Cat. No. ab92547) and CCDC80 (Bioss, Cat. No. bs-7992R).
TUNEL assay
TUNEL assay to stain the apoptotic cells of the mouse pancreatic cancer tissues was performed using DeadEnd Fluorometric TUNEL system (Promega. Cat. No. G3250) with the provided protocol. In brief, the tumour tissue sections were deparaffinised with xylene and rehydrate using graded ethanol. To permeabilize the tissues, the tissue slides were incubated with Proteinase K and then covered with equilibration buffer and rTdT buffer. SSC buffer was used to stop the reaction, and the nuclei were stained with DAPI (Vector lab, Cat. No. H-1200). Apoptotic cells (green) were detected with fluorescence microscope (100×).
Migration and invasion assays
Either vactosertib, nal-IRI/5-FU, or combination of vactosertib and nal-IRI/5-FU was added to 5 × 105 pancreatic cancer cell lines on 60 mm plates for 48 hours. Cells were detached from the plates by trypsin-EDTA (WelGENE). Approximately 5 × 104 cells were seeded in transwells for migration (Falcon, Cat. No. 353097) and invasion (Corning, Cat. No. 354480) assays and incubated for 48 hours in a 37 °C cell culture incubator. Then, we fixed the migrated and invaded cells with 70% ethanol and stained them with 0.05% crystal violet.
Colony formation assay
We seeded 5 × 102 Panc02 cells per well on a 6-well plate and incubated the plate for one week with medium changes every three days. After the cells formed colonies, we fixed the cells and stained them with methylene blue in 50% ethanol.
Western blot analysis
We followed the western blot protocol as described previously48. Briefly, PANC-1 and Panc02 cells were washed twice with PBS and lysed with RIPA buffer, 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulphate and a protease inhibitor cocktail (Sigma, P8340) on ice for 30 minutes. We measured protein concentrations using a Pierce BCA protein assay kit (Thermo Scientific, Cat. No. 23225). Proteins were heated at 95 °C in SDS sample buffer, separated by SDS-PAGE, transferred to PVDF membrane and incubated overnight with the indicated primary antibodies. The blots were detected by Amersham Imager 600 system (GE Healthcare Life Sciences, UK). The blots are cropped, and the full-length images are included in Supplementary Information.
RNA isolation, reverse transcription, and quantitative RT-PCR
RNA from cell lines and murine pancreatic tumour tissues was isolated using an easy-BLUE Total RNA extraction kit (Promega, Cat. No. 17061). Two micrograms of RNA were used for reverse transcription by M-MLV reverse transcriptase (Promega, Cat. No. M1705). Quantitative RT-PCR was performed using a QuantStudio5 Real-Time PCR instrument (Applied Biosystems) with TOPreal qPCR 2x PreMIX (Enzynomics, Cat. No. RT500M). Gene expression was normalized to Gapdh.
RNA sequencing and data analysis
The quality of purified mouse tumour RNA was measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, CA). All extractions delivered an RNA integrity number (RIN) value of 9.2–9.9 and 28 S/18 S ratio of 1.1–1.6. RNA-Seq libraries were prepared using a TruSeq RNA Sample Prep Kit according to the manufacturer’s manual (Illumina, Inc., San Diego, CA). Using 1 μg of the qualified RNA in each sample, poly(A) mRNA was enriched by magnetic beads with oligo(dT) and then fragmented into approximately 200 bp inserts by sonication. The synthesized cDNA was subjected to end-repair and poly(A) tailing and connected with sequencing adapters using a TruSeq Stranded mRNA Sample Prep Kit (Illumina, Inc., San Diego, CA). The proper cDNA fragments, purified by a BluePippin instrument (Sage Science, MA) according to the manufacturer’s instructions, were selected for further PCR amplification. The final library sizes ranged between 350–450 bp. Subsequently, the libraries were subjected to paired-end sequencing with a 100 bp read length using an Illumina HiSeq 2500 platform. The reads generated by the Illumina HiSeq 2500 platform were then processed for quality with the following criteria: low-quality reads that contained more than 10% inaccurate bases were marked as ‘N,’ and reads with >40% low quality bases (i.e., quality score < 20) were discarded. The quality scores were analysed using the fastqc tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the filtration process was performed using in-house scripts.
Bioinformatics analysis
Clean reads were aligned to the mouse reference genome (Ensembl 84) using TopHat49 with a set of its gene model annotations. Gene expression was quantified using Cufflinks50. Differential expression (DE) analysis between sample groups of interest was performed using Cuffdiff50 with cut-offs set at P < 0.05 and ≥1.5-fold change. In addition, because of possible noise at very low levels of expression, genes expressed at ≥1 FPKM in at least one sample were retained for DE analysis. Hierarchical clustering for selected genes was conducted with MeV (http://mev.tm4.org) using the Euclidean distance and complete linkage method. Functional annotation for module members was performed using DAVID51, and relevant gene ontology (GO) terms were selected with a cut-off of P < 0.001. Gene set enrichment analysis (GSEA) was further performed to examine the critical GOs of the transcriptome52. For this, the estimated expression levels of all genes were run through GSEA, and then the enrichment scores were calculated according to the rank-ordered gene list. Significance scores were computed by 1,000 nonparametric permutation tests.
Statistics
All the data were expressed as the mean with SD. Statistical significance between all quantitative data are analyzed by Student t-tests and one-way ANOVA using GraphPad Prism version 5 (GraphPad Software Inc.).
Data availability
The RNA-Seq data is available in the NCBI Sequence Read Archive (SRA) database under the accession number SRP229044 (https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP229044&o=acc_s%3Aa).
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2018. CA: a cancer journal for clinicians. 68, 7–30 (2018).
Ryan, D. P., Hong, T. S. & Bardeesy, N. Pancreatic Adenocarcinoma. New England Journal of Medicine. 371, 1039–1049 (2014).
Zhou, B. et al. Early detection of pancreatic cancer: Where are we now and where are we going? International Journal of Cancer. 141, 231–241 (2017).
Kleeff, J. et al. Pancreatic cancer. Nature reviews. Disease primers. 2, 16022 (2016).
Notta, F., Hahn, S. A. & Real, F. X. A genetic roadmap of pancreatic cancer: still evolving. Gut. 66, 2170–2178 (2017).
Lomberk, G. et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nature Communications. 9, 1978 (2018).
DeVita, V. T. Jr. & Chu, E. A history of cancer chemotherapy. Cancer research. 68, 8643–8653 (2008).
Burris, H. A. III et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 15, 2403–2413 (1997).
Conroy, T. et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. The New England journal of medicine. 364, 1817–1825 (2011).
Von Hoff, D. D. et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. The New England journal of medicine. 369, 1691–1703 (2013).
Drummond, D. C. et al. Development of a highly active nanoliposomal irinotecan using a novel intraliposomal stabilization strategy. Cancer research. 66, 3271–3277 (2006).
Ko, A. H. et al. A multinational phase 2 study of nanoliposomal irinotecan sucrosofate (PEP02, MM-398) for patients with gemcitabine-refractory metastatic pancreatic cancer. British journal of cancer. 109, 920–925 (2013).
Wang-Gillam, A. et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): a global, randomised, open-label, phase 3 trial. Lancet. 387, 545–557 (2016).
Massague, J. TGFbeta in. Cancer. Cell. 134, 215–230 (2008).
Padua, D. & Massagué, J. Roles of TGFβ in metastasis. Cell Research. 19, 89–102 (2009).
Melisi, D. et al. LY2109761, a novel transforming growth factor beta receptor type I and type II dual inhibitor, as a therapeutic approach to suppressing pancreatic cancer metastasis. Molecular cancer therapeutics. 7, 829–840 (2008).
Gadir, N., Jackson, D. N., Lee, E. & Foster, D. A. Defective TGF-β signaling sensitizes human cancer cells to rapamycin. Oncogene. 27, 1055 (2007).
Keedy, V. L. et al. Association of TGF-β responsive signature with anti-tumor effect of vactosertib, a potent, oral TGF-β receptor type I (TGFBRI) inhibitor in patients with advanced solid tumors. Journal of Clinical Oncology. 36, 3031–3031 (2018).
Jung, S. Y. et al. Pharmacokinetic characteristics of vactosertib, a new activin receptor-like kinase 5 inhibitor, in patients with advanced solid tumors in a first-in-human phase 1 study. Invest New Drugs. (2019).
Son, J. Y. et al. EW-7197, a novel ALK-5 kinase inhibitor, potently inhibits breast to lung metastasis. Molecular cancer therapeutics. 13, 1704–1716 (2014).
Yoon, J. H. et al. Activin receptor-like kinase5 inhibition suppresses mouse melanoma by ubiquitin degradation of Smad4, thereby derepressing eomesodermin in cytotoxic T lymphocytes. EMBO molecular medicine. 5, 1720–1739 (2013).
Park, S. et al. Identification of Epithelial-Mesenchymal Transition-related Target Genes Induced by the Mutation of Smad3 Linker Phosphorylation. J Cancer Prev. 23, 1–9 (2018).
Pei, G. et al. The function of FAK/CCDC80/E-cadherin pathway in the regulation of B16F10 cell migration. Oncology letters. 16, 4761–4767 (2018).
Ferraro, A. et al. Tumor suppressor role of the CL2/DRO1/CCDC80 gene in thyroid carcinogenesis. The Journal of clinical endocrinology and metabolism. 98, 2834–2843 (2013).
Bommer, G. T. et al. DRO1, a gene down-regulated by oncogenes, mediates growth inhibition in colon and pancreatic cancer cells. The Journal of biological chemistry. 280, 7962–7975 (2005).
Shen, W. et al. TGF-beta in pancreatic cancer initiation and progression: two sides of the same coin. Cell & bioscience. 7, 39 (2017).
Hong, S. M., Park, J. Y., Hruban, R. H. & Goggins, M. Molecular signatures of pancreatic cancer. Arch Pathol Lab Med. 135, 716–727 (2011).
Bartsch, D. et al. Higher frequency of DPC4/Smad4 alterations in pancreatic cancer cell lines than in primary pancreatic adenocarcinomas. Cancer Lett. 139, 43–49 (1999).
Ahmed, S. et al. The TGF-beta/Smad4 Signaling Pathway in Pancreatic Carcinogenesis and Its Clinical Significance. Journal of clinical medicine. 6, (2017).
Miyazono, K. et al. Intracellular and extracellular TGF-beta signaling in cancer: some recent topics. Front Med. 12, 387–411 (2018).
Madar, S., Goldstein, I. & Rotter, V. ‘Cancer associated fibroblasts’–more than meets the eye. Trends Mol Med. 19, 447–453 (2013).
Sannino, G. et al. Role of BCL9L in transforming growth factor-beta (TGF-beta)-induced epithelial-to-mesenchymal-transition (EMT) and metastasis of pancreatic cancer. Oncotarget. 7, 73725–73738 (2016).
Shao, T. et al. Gamma synuclein is a novel Twist1 target that promotes TGF-beta-induced cancer cell migration and invasion. Cell Death Dis. 9, 625 (2018).
Hallas, C. et al. BCL9L expression in pancreatic neoplasia with a focus on SPN: a possible explanation for the enigma of the benign neoplasia. BMC cancer. 16, 648 (2016).
Liu, C. et al. Synuclein gamma predicts poor clinical outcome in colon cancer with normal levels of carcinoembryonic antigen. BMC cancer. 10, 359 (2010).
Cirak, Y. et al. Predictive and prognostic values of BubR1 and synuclein-gamma expression in breast cancer. Int J Clin Exp Pathol. 8, 5345–5353 (2015).
Li, Z. et al. Overexpression of synuclein-gamma in pancreatic adenocarcinoma. Cancer. 101, 58–65 (2004).
Nguyen, T. M. et al. Loss of ITM2A, a novel tumor suppressor of ovarian cancer through G2/M cell cycle arrest, is a poor prognostic factor of epithelial ovarian cancer. Gynecologic oncology. 140, 545–553 (2016).
Schafer, S. A. et al. Correlation between SFRP1 expression and clinicopathological parameters in patients with triple-negative breast cancer. Future oncology. 15, 1921–1938 (2019).
Kumar, A., Gosipatala, S. B., Pandey, A. & Singh, P. Prognostic Relevance of SFRP1 Gene Promoter Methylation in Colorectal Carcinoma. Asian Pac J Cancer Prev. 20, 1571–1577 (2019).
Garcia-Tobilla, P. et al. SFRP1 repression in prostate cancer is triggered by two different epigenetic mechanisms. Gene. 593, 292–301 (2016).
Davaadorj, M. et al. Loss of SFRP1 Expression Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Anticancer Res. 36, 659–664 (2016).
Ferragud, J. et al. Transcriptional repression of the tumor suppressor DRO1 by AIB1. FEBS letters. 585, 3041–3046 (2011).
Tremblay, F. et al. Bidirectional modulation of adipogenesis by the secreted protein Ccdc80/DRO1/URB. The Journal of biological chemistry. 284, 8136–8147 (2009).
Grill, J. I. et al. Loss of DRO1/CCDC80 results in obesity and promotes adipocyte differentiation. Molecular and cellular endocrinology. 439, 286–296 (2017).
Herbst, A., Bayer, C., Wypior, C. & Kolligs, F. T. DRO1 sensitizes colorectal cancer cells to receptor-mediated apoptosis. Oncology letters. 2, 981–984 (2011).
Grill, J. I. et al. Dro1/Ccdc80 inactivation promotes AOM/DSS-induced colorectal carcinogenesis and aggravates colitis by DSS in mice. Carcinogenesis. 39, 1176–1184 (2018).
Kang, J. M. et al. KIAA1324 Suppresses Gastric Cancer Progression by Inhibiting the Oncoprotein GRP78. Cancer research. 75, 3087–3097 (2015).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology. 14, R36 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7, 562–578 (2012).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4, 44–57 (2009).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 102, 15545–15550 (2005).
Acknowledgements
This study was supported by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI14C2640). We want to thank Yealeen Jeong for editing and proofreading this manuscript.
Author information
Authors and Affiliations
Contributions
E.H., S.P., S.J.K., A.O., J.P., S.H.P. and J.O.P. designed the experiments. E.H., J.S.H., S.L., H.A., and J.M.K. performed the experiments. A.O. performed histological analysis, and C.H. performed RNA-sequencing analysis. E.H. and S.P. drafted the manuscript, and J.P., A.O., J.O.P. and S.J.K. revised the manuscript. All of the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
S.J.K. is a founder, board member, and shareholder of Medpacto Inc. and a shareholder of TheragenEtex. C.P.H. is a paid employee of TheragenEtex. S.H.P. is a shareholder of Medpacto Inc. E.H., S.P., A.O., J.S.H., S.L., H.A., and J.O.P. declare that they have no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hong, E., Park, S., Ooshima, A. et al. Inhibition of TGF-β signalling in combination with nal-IRI plus 5-Fluorouracil/Leucovorin suppresses invasion and prolongs survival in pancreatic tumour mouse models. Sci Rep 10, 2935 (2020). https://doi.org/10.1038/s41598-020-59893-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59893-5
This article is cited by
-
Combination treatment of T1-44, a PRMT5 inhibitor with Vactosertib, an inhibitor of TGF-β signaling, inhibits invasion and prolongs survival in a mouse model of pancreatic tumors
Cell Death & Disease (2023)
-
Chemotherapy-induced metastasis: molecular mechanisms and clinical therapies
Acta Pharmacologica Sinica (2023)
-
Targeting TGF-β signal transduction for fibrosis and cancer therapy
Molecular Cancer (2022)
-
Mutations in key driver genes of pancreatic cancer: molecularly targeted therapies and other clinical implications
Acta Pharmacologica Sinica (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.