Abstract
Citrus species are among the most economically important fruit crops. Physiological characteristics and molecular mechanisms associated with de-etiolation have been partially revealed. However, little is known about the mechanisms controlling the expression and function of genes associated with photosynthesis and chlorophyll biosynthesis in variegated citrus seedlings. The lower biomass, chlorophyll contents, and photosynthetic parameter values recorded for the variegated seedlings suggested that chlorophyll biosynthesis was partially inhibited. Additionally, roots of the variegated seedlings were longer than the roots of green seedlings. We obtained 567.07 million clean reads and 85.05 Gb of RNA-sequencing data, with more than 94.19% of the reads having a quality score of Q30 (sequencing error rate = 0.1%). Furthermore, we detected 4,786 and 7,007 differentially expressed genes (DEGs) between variegated and green Shiranuhi and Huangguogan seedlings. Thirty common pathways were differentially regulated, including pathways related to photosynthesis (GO: 0015979) and the chloroplast (GO: 0009507). Photosynthesis (44 and 63 DEGs), photosynthesis-antenna proteins (14 and 29 DEGs), and flavonoid biosynthesis (16 and 29 DEGs) pathways were the most common KEGG pathways detected in two analyzed libraries. Differences in the expression patterns of PsbQ, PetF, PetB, PsaA, PsaN, PsbP, PsaF, Cluster-2274.8338 (ZIP1), Cluster-2274.38688 (PTC52), and Cluster-2274.78784 might be responsible for the variegation in citrus seedlings. We completed a physiological- and transcriptome-level comparison of the Shiranuhi and Huangguogan cultivars that differ in terms of seedling variegation. We performed mRNA-seq analyses of variegated and green Shiranuhi and Huangguogan seedlings to explore the genes and regulatory pathways involved in the inhibition of chlorophyll biosynthesis and decreases in Chl a and Chl b contents. The candidate genes described herein should be investigated in greater detail to further characterize variegated citrus seedlings.
Similar content being viewed by others
Introduction
Citrus, which have been cultivated globally, is one of the most important fruit species1. Shiranuhi [Citrus reticulata × (Citrus reticulata × Citrus sinensis)] and Huangguogan (Citrus reticulata × Citrus sinensis) are relatively new hybrid citrus cultivars grown in China2. Etiolation, which occurs in many angiosperms, refers to the phenomenon that leaves appear yellow under dark conditions. Seedlings undergo skotomorphogenesis in lightless culture condition, and leaves take on the color of carotenoids. This phenomenon results in a rapid elongation of the hypocotyl3. Changes in plant morphology and growth are the ultimate reflection of plant damages caused by etiolation. This phenomenon leads to decreased leaf and optical areas, the production of dwarf plants, a weakened growth potential, and even death. A stable supply of chlorophyll, which is the main photosynthetic pigment of plants, is a key requirement for the normal development of photosynthesis4. Analyses of light regulation5, ethylene responses6, riboflavin biosynthesis7, endogenous abscisic acid levels3, phospholipid hydroperoxide glutathione peroxidase activity8, and proteomics analysis9 have been used to examine plant growth and development induced by etiolation. Many studies have focused on the expression analysis of related genes during photosynthesis in greening, and the regulation of the corresponding protein levels10. However, there have been relatively few transcriptome-level studies of variegation in citrus seedlings.
Variegated plants, which have both green and white or yellow areas on the same leaf, are considered as the invaluable materials for studying chloroplast biosynthesis, development and maintenance11. The white sectors of var2, which is one of the Arabidopsis (Arabidopsis thaliana) mutants showing leaf variegation, are active tissues that are formed by viable cells with undifferentiated plastids12. Chloroplast development and chlorophyll accumulation are inhibited at the albinism growth stage of albino tea cultivars13. In photosynthetic organisms, chlorophyll and carotenoid are the main pigments of plants that capture light energy. Earlier studies revealed that the etiolated growth of seedlings or leaves considerably affects chloroplast development and chlorophyll metabolism14,15,16. Whether the seedlings undergo etiolation or exhibit variegation, both lead to color changes of leaves.
Next-generation sequencing (NGS) technology is a rapid and cost-effective approach to analyzing a large number of protein-coding genes17,18. This technology can be used to address questions related to ecological comparative and evolutionary genomics in non-model organisms19. Based on the similar expression pattern, using transcriptomic data to group differentially expressed genes (DEGs) is one of the most effective methods to explore the relationships and predict functions of candidate genes20,21. Similarly expressed genes are often functionally related22. In the present study, we identified variegation -related genes and examined the pathways associated with photosynthesis and chlorophyll biosynthesis using RNA-sequencing (RNA-seq) combined with the analysis of chlorophyll contents to characterize the variegation in citrus seedlings. Our findings may help clarify the molecular basis of variegation.
We recently analyzed the transcriptome of etiolated citrus seedlings, there were variegated and green seedlings in both Shiranuhi and Huangguogan under the same conditions of seedling germination23. Although variegated plants exist widely in nature, the mechanism of citrus leaf variegation and the mechanisms regulating the expression patterns of well-characterized genes involved in photosynthesis and chlorophyll biosynthesis are still unknown. In order to study the color difference of citrus leaf after germination under the same culture condition, transcriptome sequencing was performed on the leaves of citrus seedlings. In the present study, we compared the leaf biomass and chlorophyll contents of variegated and green Shiranuhi and Huangguogan seedlings. In addition to the differentially expressed photosynthesis-related genes, the expression profiles of chlorophyll biosynthesis-related genes were analyzed to reveal the molecular mechanism underlying variegation.
Results
Biomass accumulation of variegated and green seedlings
Variegated and green Shiranuhi and Huangguogan seedlings were sampled at 20 days after germination to measure dry weight, length, and chlorophyll contents. We observed a significant difference in shoot and root dry weight between the variegated and green seedlings. Additionally, the roots of variegated seedlings were longer than those of green seedlings for both analyzed cultivars, while the opposite trend was observed for the shoots. Furthermore, the shoot lengths of green seedlings were longer than those of variegated seedlings in both Shiranuhi and Huangguogan, while the roots were shorter (Fig. 1). These results indicated that the variegation of citrus leaves had different effects on seedling root and shoot lengths.
Chlorophyll content and photosynthesis of variegated seedlings
The chlorophyll a (Chl a) concentration of variegated Shiranuhi and Huangguogan seedlings were 0.56 and 0.71 mg g−1 fresh weight, respectively (Fig. 2), while the corresponding concentrations in green seedlings were 1.86 and 2.22 mg g−1 fresh weight, respectively. Similarly, chlorophyll b (Chl b) and carotenoid contents as well as the composition and proportion of photosynthetic pigments exhibited the same trends as the Chl a content.
The photosynthetic parameters differed between the Huangguogan and Shiranuhi variegated and green seedlings (Fig. 3). For example, the net photosynthetic rate (Pn), intercellular CO2 concentration (Ci), and transpiration rate (Tr) were lower in variegated seedlings than in green seedlings. Additionally, the photosynthetic parameter values were lower for Huangguogan than for Shiranuhi seedlings, except for the Ci.
Sequencing and assembly of expressed genes
To investigate the genome-wide expression patterns of variegated seedlings, more than 10 leaves harvested from each seedling type for each cultivar at 20 days after germination were analyzed by RNA-seq. We constructed four cDNA libraries, namely R_V and R_G, which represent variegated and green Shiranuhi seedlings, respectively, and Y_V and Y_G, which correspond to variegated and green Huangguogan seedlings, respectively. After removing the sequencing adapters and low-quality data, we obtained 567.07 million clean reads and 85.05 Gb of RNA-seq data. More than 94.19% of the reads had a quality score of Q30 (sequencing error rate = 0.1%). The sequencing data statistics are listed in Table 1. The raw data were deposited in the NCBI Gene Expression Omnibus database (accession number GSE90935).
Evaluation of gene expression levels
The number of fragments per kilobase of exon per million fragments mapped (FPKM) was used to quantify the unigene expression levels, which were detected by RNA-seq with high sensitivity. Hierarchical cluster analyses were conducted with 66 DEGs identified for variegated and green Shiranuhi and Huangguogan seedlings. Genes with the same or similar expression profiles were clustered to present the gene sets exhibiting different expression patterns under various experimental conditions (Fig. 4).
Identification and functional annotation of differentially expressed genes
During the screening of DEGs, we used P value < 0.00524 and |log2 (fold change)| > 1 as the threshold criteria to determine the significance of the differences in gene expression levels. A total of 4,786 and 7,007 DEGs between variegated and green Shiranuhi and Huangguogan seedlings were classified into the gene ontology (GO) categories biological process (GO-BP), molecular function (GO-MF), and cellular component (GO-CC) (Figs 5–6). For the R_V library, significant differences in the enrichment of 30 biological processes were detected in the GO-BP class, including photosynthesis (79 DEGs), cellular hormone metabolic process (44 DEGs), hormone metabolic process (44 DEGs), regulation of hormone levels (44 DEGs), oxidation-reduction process (485 DEGs), and other processes. Additionally, ADP binding (79 DEGs), oxidoreductase activity (482 DEGs), 3-beta-hydroxy-delta 5-steroid dehydrogenase activity (39 DEGs), and 19 other processes were significantly enriched in the GO-MF class. Furthermore, photosystem (72 DEGs), photosystem II oxygen-evolving complex (25 DEGs), chloroplast (40 DEGs), chloroplast thylakoid membrane (14 DEGs), chloroplast thylakoid (14 DEGs), photosystem II (51 DEGs), and photosystem I (22 DEGs) were significantly enriched in the GO-CC class (Fig. 6). For the Y_V library, significant differences in enrichment were observed for 83 sub-categories in the GO-BP class, 28 in the GO-MF class, and 32 in the GO-CC class. A comparison of both libraries indicated that 15 sub-categories were related to the GO-BP class, such as oxidation-reduction process (GO: 0055114), single-organism metabolic process (GO: 0044710), and photosynthesis (GO: 0015979). Meanwhile, 13 sub-categories were related to the GO-CC class, such as photosystem I (GO: 0009522), chloroplast thylakoid (GO: 0009534), chloroplast thylakoid membrane (GO: 0009535), photosynthetic membrane (GO: 0034357), photosystem (GO: 0009521), and chloroplast (GO: 0009507). In contrast, only oxidoreductase activity (GO: 0016491) was related to the GO-MF class (Fig. 6).
Differential expression analysis of unigenes in the variegated seedlings (M) versus the green seedlings (G) of Shiranuhi (R) and Huangguogan (Y). The log2(fold change) is used as the x-axis and −log10(pdaj) is used as the y-axis. 2273 and 3880 unigenes were up-regulated in R_V and Y_V (highlighted in green), and 2513 and 3127 unigenes were down-regulated (highlighted in red), respectively.
To investigate the biological pathways important for the variegation in Shiranuhi and Huangguogan seedlings, the DEGs between variegated and green seedlings were further annotated based on the reference pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Fig. 7). These DEGs in the R_V and Y_V libraries were assigned to 117 and 119 KEGG pathways, respectively. The pathway search results were sorted based on the number of hits, and photosynthesis (44 and 63 DEGs), photosynthesis-antenna proteins (14 and 29 DEGs), and flavonoid biosynthesis (16 and 29 DEGs) were the most highly represented pathways in both libraries. Genes associated with cysteine and methionine metabolism (47 DEGs), monobactam biosynthesis (9 DEGs), and ABC transporters (15 DEGs) were also common in the R_V library (Fig. 7). In contrast, genes related to carbon fixation in photosynthetic organisms (80 DEGs), ribosome (154 DEGs), and porphyrin and chlorophyll metabolism (41 DEGs) were common in the Y_V library (P value < 0.05).
Analysis of chlorophyll biosynthesis and photosynthesis-related transcripts
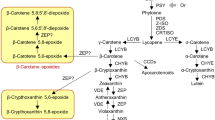
Chlorophyll is a key photosynthetic pigment in plant chloroplasts, and chlorophyll metabolism is an important factor influencing crop yield25. To investigate whether photosynthesis pathway genes are involved in this decrease, we analyzed the expression patterns of genes encoding regulatory enzymes associated with chlorophyll biosynthesis (Fig. 8). Fifteen enzymes are required for the chlorophyll biosynthesis from glutamyl-tRNA to Chl b26. Magnesium (Mg) chelation is the first step in the cholorophyll branch of porphyin metabolism, forming magnesium-protoporphyrin (MgP, EC: 2.1.1.11), which is followed by a SAM-dependent methylation of the 6-propionate side chain to form Mg-protoporphyrin monomethyl ester (MgPME)27. We observed that the expression of the gene encoding magnesium chelatase subunit I (MgCE I, EC: 6.6.1.1) was down-regulated from 580.67 to 15.05 FPKM (Cluster-2274.70486) in the Y_V library. Magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase (MgPMEC, EC: 1.14.13.81) is another key enzyme involved in chlorophyll biosynthesis. Our RNA-seq data revealed a considerable difference in the abundance of MgPMEC transcripts (Cluster-2274.8338), with 46.77 FPKM in the Y_G library and 0 FPKM in the Y_V library, this was consistent with previous research about variegated Epipremnum aureum11. Meanwhile, protochlorophyllide reductase (POR, EC: 1.3.1.33) catalyzes a reaction with divinyl-protochlorophyllide as a substrate to synthesize divinyl-chlorophyllide a25. The expression levels of the gene encoding POR (Cluster-2274.3601) were down-regulated, with 19.73 FPKM in the Y_G library and 0 FPKM in the Y_V library. Furthermore, in the chlorophyll biosynthesis pathway, 7-hydroxymethyl chlorophyll a reductase (HCAR, EC: 1.17.7.2) converts Chl b to 7-hydroxymethyl Chl a and ultimately to Chl a28. We observed no changes in the expression levels of the gene encoding HCAR in the Y_V library, but it was down-regulated from 51.14 to 10.30 FPKM (Cluster-2274.20724) in the R_V library.
Chlorophyll metabolism-related gene expression-level changes in the chlorophyll biosynthesis pathways of (A) Shiranuhi and (B) Huangguogan seedlings. Numbers in boxes correspond to the following EC numbers: 4.2.1.24, porphobilinogen synthase. 2.5.1.61, hydroxymethylbilane synthase. 4.1.1.37, uroporphyrinogen decarboxylase. 4.2.1.75, uroporphyrinogen-III synthase. 4.1.1.37, uroporphyrinogen decarboxylase. 1.3.98.3, oxygen-independent coproporphyrinogen III oxidase. 1.3.3.4, protoporphyrinogen oxidase. 6.6.1.1, magnesium chelatase subunit H, D, I. 2.1.1.11, magnesium-protoporphyrin O-methyltransferase. 1.14.13.81, magnesium- protoporphyrin IX monomethyl ester (oxidative) cyclase. 1.3.1.75, divinyl chlorophyllide a 8-vinyl-reductase. 1.3.1.33, protochlorophyllide reductase. 2.5.1.62, chlorophyll synthase. 3.1.1.14, chlorophyllase. 1.14.13.122, chlorophyllide a oxygenase. 1.1.1.294, chlorophyll(ide) b reductase. 1.17.7.2, 7-hydroxymethyl chlorophyll a reductase.
In comparisons between the R_V and R_G libraries and between the Y_V and Y_G libraries, we detected six common DEGs related to porphyrin and chlorophyll metabolism, namely Cluster-2274.78784 (K08099, 3.1.1.14), Cluster-2274.38688 (K13071, 1.14.15.17), Cluster-2274.8338 (K04035, 1.14.13.81), Cluster-2274.66782 (K03403, 6.6.1.1), Cluster-2274.56424 (K13071, 1.14.15.17), and Cluster-2274.70486 (K03428, 2.1.1.11) (Fig. 8). The comparison between the R_V and R_G libraries revealed 44 DEGs related to the photosynthesis pathway, including 13 genes related to photosystem I (PSI), 15 related to photosystem II (PSII), four related to the cytochrome b6/f complex, three related to photosynthetic electron transport, and nine related to the F-type ATPase. In contrast, the comparison between the Y_V and Y_G libraries detected 63 DEGs associated with the photosynthesis pathway, including 26, 15, 5, 10, and 7 related to PSI, PSII, cytochrome b6/f complex, photosynthetic electron transport, and F-type ATPase, respectively.
In comparisons between the R_V and Y_V libraries and between the R_G and Y_G libraries, the following nine common DEGs associated with the photosynthesis pathway were identified: Cluster-2274.72645 (PsbQ), Cluster-2274.39503 (PetF), Cluster-2274.81464 (PetF), Cluster-2274.51835 (PetB), Cluster-2274.57179 (F-type ATPase a), Cluster-2274.57012 (PsaA), Cluster-19856.1 (PsaN), Cluster-17851.0 (PsbP), and Cluster-2274.55964 (PsaF), and these DEGs were marked on KEGG imagery (map00195) from Kanehisa laboratories29,30 (Fig. 9). Additionally, 15 genes involved in photosynthetic activities and chlorophyll biosynthesis may be associated with the differences in leaf color between the R_V and Y_V seedlings (Figs 8,9).
Differentially expressed genes mapped to photosynthesis pathway. The known pathways were obtained from the KEGG database. Red square denotes up-regulated expression in variegated seedlings of Shiranuhi and Huangguogan compared with that of green seedlings, respectively, while green square denotes both up- and down-regulated genes. We have got the formal permission of KEGG imagery (map00195)29,30 from Kanehisa laboratories.
qRT-PCR validation
We selected photosynthetic genes (PsbQ, PetF, PetB, PsaA, PsaN, PsbP, and PsaF) and 6 porphyrin and chlorophyll metabolism pathway genes from common DEGs in R_V and Y_V libraries and evaluated their expression profiles using quantitative real-time PCR. Actin (GenBank: XM 006480741.2) was selected for internal controls. The verification results of the 7 photosynthesis-related and 6 chlorophyll metabolism-related DEGs demonstrated that nearly all of the common photosynthetic genes showed similar expression patterns of RNA-seq analysis (Fig. 10). In the porphyrin and chlorophyll metabolism pathway, there were three common DEGs (Cluster-2274.8338, Cluster-2274.38688, Cluster-2274.78784) with a similar expression pattern in R_V and Y_V seedlings. The other three common DEGs showed the opposite expression pattern, up-regulated in R_V and down-regulated in Y_V (Fig. 10).
Expression pattern of 7 photosynthesis-related and 6 chlorophyll metabolism-related genes as obtained by RNA-seq and qRT-PCR. (A) qRT-PCR validation for the 7 photosynthesis-related genes of Shiranuhi. (B) qRT-PCR validation for the 7 photosynthesis-related genes of Huangguogan. (C) qRT-PCR validation for the 6 chlorophyll metabolism-related genes of Shiranuhi. (D) qRT-PCR validation for the 6 chlorophyll metabolism-related genes of Huangguogan. R_V, R_G, Y_V and Y_G represent normalized expression levels for the DEGs in the Shiranuhi and Huangguogan libraries, respectively. FC is the ratio of FPKM between variegated and green seedlings.
Discussion
The purpose of this research was to study the differences in the transcriptome expression of variegated and green Huangguogan and Shiranuhi seedlings, especially regarding photosynthesis and chlorophyll biosynthesis. We identified major transcriptomic features in the variegated seedlings.
Photosynthetic activities are affected by stomatal conductance (Gs), Ci, Tr, and chlorophyll fluorescence31. The Pn directly reflects the photosynthetic capacity32, while Ci is an important indicator of stomatal activity. During photosynthesis, a decrease in Tr caused by a decrease in Gs leads to a significant decrease in transpiration, which can inhibit the absorption and transport of water and nutrients33. In addition to shorter roots, the variegated seedlings also had significantly lower seedling biomass, chlorophyll content, and photosynthetic parameter values (P < 0.05) compared with the green seedlings (Figs 1–3). The identification and functional annotation of DEGs indicated that the photosynthesis pathway genes (44 and 63 DEGs) were the most common genes in the R_V and Y_V libraries (based on the number of hits for DEGs), with the lowest corrected P values (8.17212942029e-11 and 6.07238386484e-12) (Fig. 7).
Transcriptional regulation is important for coordinating the conversion of glutamyl-tRNA to the final product (Chl a and Chl b). There are 15 enzymes encoded by 27 genes in Arabidopsis thaliana in chlorophyll synthesis4. The six-electron oxidation of protogen IX is catalyzed to protoporphyrin IX (proto IX) by Protoporphyrinogen IX (protogen IX) oxidase (PPOX), with flavin as the cofactor34. Previous studies revealed that MgPMT (Mg-protoporphyrin IX methyltransferase) activity is inversely related to the transcriptional activities associated with Mg-chelatase and ALA synthesis, and restricts the formation of chlorophyll4,35. Low expression levels of Cluster-2274.8338 (K04035, 1.14.13.81) lead to a decrease in divinyl protochlorophyllide synthesis, while the up-regulated expression of Cluster-2274.78784 (K08099, 3.1.1.14) induces the transformation of Chl a to Chlorophyllide a. Furthermore, the up-regulated expression of the Cluster-2274.56424 (K13071, 1.14.15.17) gene enhances the synthesis of the chlorophyll catabolite during chlorophyll biosynthesis.
Three common differentially expressed genes of porphyrin and chlorophyll metabolism pathway with the same expression pattern (Cluster-2274.8338, Cluster-2274.38688, Cluster-2274.78784) in the variegated Shiranuhi and Huangguogan seedlings were blasted in Citrus Sinensis Annotation Project (http://citrus.hzau.edu.cn/orange/index.php). The sequence of Cluster-2274.8338 was similar to Cs6g16200 (the e-value was 4e-77, and ident was 83%), which is the ZIP1-like gen of Citrus sinensis. In addition, the sequences of ZIP1 (GenBank: FJ940751.1) of Oryza sativa Japonica Group (Japanese rice) in NCBI have been compared with transcriptome data by sequence alignment. The percentage of identical matches between ZIP1 and Cluster-2274.8338 was 99.92%, the total score was 2261, and the expect value (e-value) was 1.37e-26. On the other hand, Cluster-2274.8338 (ZIP1) was down-regulated in R_V and Y_V, and the log2 FC were −2.7399 and −2.5467, respectively (Fig. 10). Previous studies have found that EaZIP transcripts may not accumulate or become unstable in the early stage of plastid development36, leading to the loss of MPE cyclase, and the low expression results in chlorophyll losing and forming the diverse phenotype of leaf color11. Cluster-2274.38688 was similar to Cs8g15480.1, which encodes protochlorophyllide-dependent translocon component 52 (PTC52). PTC52 is considered to be part of a unique transposon and is most abundant in etiolated plants37. In this study, Cluster-2274.38688 gene was found to be up-regulated in variegated seedlings (Fig. 10). In addition, Cluster-2274.78784 was similar to Cs9g07520 (the total score was 3675, query cover was 78%, e-value was 0, and ident was 99%), which was noted to be involved in chlorophyll catabolic process (GO: 0015996). The up-regulated expression of Cluster-2274.78784 might lead to the decomposition of Chl b to chlorophyllide b. These results indicated that the formation of variegated seedlings may be caused by decreased chlorophyll synthesis and increased catabolism.
Nuclear-encoded genes affecting photosynthesis are co-expressed to assemble PSI and PSII during the de-etiolation of plants34,38. The transcription levels of several photosynthesis-related genes, such as PsbA39, LHCA1–6 and LHCB1–640, as well as LSU and SSU, are regulated by light during chloroplast biogenesis in de-etiolating plants41. In this study, we detected 9 common DEGs involved in the photosynthesis pathways of the variegated Shiranuhi and Huangguogan seedlings. Additionally, we identified 6 common DEGs related to porphyrin and chlorophyll metabolism. The differential expression of these genes may be critical for the leaves variegation in citrus seedlings, but need to be confirmed in future work.
Materials and Methods
Plant materials
The Shiranuhi and Huangguogan hybrid citrus cultivars analyzed in this study (Fig. 11) were provided by the Institute of Pomology and Olericulture, Sichuan Agricultural University, China. Refer to the method of Xiong et al.23 for seed germination and seedling culture. Seeds were presoaked in water for 4 h and then incubated at 25 °C for 3 days, sown in pots (vermiculite: perlite = 1:1) and then transferred to a growth chamber at 25 °C, 50–60% relative humidity, and 12-h light/12-h dark period. After germination (i.e., radicle breaks through the seed coat), seedlings were watered every 2 days. The Shiranuhi variegated seedlings (R_V), Shiranuhi green seedlings (R_G), Huangguogan variegated seedlings (Y_V), and Huangguogan green seedlings (Y_G) sample contained more than 20 seedlings were used for physiological and transcriptome analyses.
Photosynthetic characteristics
Three seedlings of each sample were selected as test materials. The photosynthetic parameters of the second fully unfolded leaves from the base of the seedlings were recorded 20 days after germination. The Pn, Tr, Ci, and Gs were measured using the LI-6400 portable photosynthesis system (Li-Cor, Lincoln, NE, USA). The photosynthetically active radiation and CO2 concentration were 1,200 µmol m−2 s−1 and 400 µmol m−2 s−1, respectively. Additionally, Gs and Ci were determined at a saturated light intensity of 1,000 µmol m−2 s−1 and 70% relative humidity42.
Chlorophyll and carotenoid contents
All leaves, used for the determination of photosynthetic characteristics, were harvested from variegated and green seedlings for each cultivar at 20 days after germination. Once harvested, plant materials were quick frozen using liquid nitrogen and stored at −80 °C until using. The chlorophyll and carotenoid contents of seedlings 20 days after germination were estimated as previously described using the following epuations43: Chl a = (12.7OD663 − 2.69OD645) × 10/1000 W; Chl b = (22.9OD645 − 4.68OD663) × 10/1000 W; Carotenoid = (1000OD470 − 3.27 Chl a − 104 Chl b)/229 × 10/1000 W.
Root and shoot dry weights and lengths
All seedlings, used for the determination of photosynthetic characteristics, were collected and divided into shoots and roots. The root and shoot lengths were measured as previously described44. After the green removing process at 110 °C for 24 h, the dry weight of root and shoot was measured.
RNA extraction and qualification
Three biological replicates were collected for Shiranuhi and Huangguogan seedlings. Total RNA was extracted from R_V, R_G, Y_V, and Y_G with the PureLink Plant RNA Reagent (Invitrogen, CA, USA). The purity of the extracted RNA was checked using the NanoPhotometer spectrophotometer (IMPLEN, CA, USA), while RNA integrity was assessed using the RNA 6000 Nano Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA)45.
Preparation of cDNA libraries and transcriptome sequencing
We purified mRNA from 1.5 µg total RNA using poly-T oligo-attached magnetic beads. The RNA was fragmented using divalent cations in 5 × NEBNext First Strand Synthesis Reaction Buffer. First-strand cDNA was synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H−). Second-strand cDNA was subsequently synthesized using DNA Polymerase I and RNase H23. The remaining overhangs were converted into blunt ends by exonuclease/polymerase activities. The NEBNext adapter with a hairpin loop structure was ligated to adenylated 3′ ends of DNA fragments for a subsequent hybridization. To preferentially select cDNA fragments that were 150–200 bp long, the library fragments were purified with the AMPure XP system (Beckman Coulter, Beverly, USA). The size-selected, adapter-ligated cDNA sequences were treated with 3 µl USER Enzyme (NEB, USA) at 37 °C for 15 min followed by a 5-min incubation at 95 °C. A PCR was then completed using Phusion High-Fidelity DNA polymerase, Universal PCR primers, and an Index (X) Primer. The PCR products were purified using the AMPure XP system, and library quality was assessed using the Bioanalyzer 2100 system46. The index-coded samples were clustered with the cBot Cluster Generation System of the TruSeq PE Cluster Kit v3-cBot-HS (Illumina)47. The resulting libraries were sequenced using the HiSeq. 4000 system (Illumina), which generated paired-end reads.
Quality control
Refer to the previous method, raw data (raw reads) in a fastq format were first processed using in-house perl scripts. Clean data (clean reads) were obtained by removing reads containing the adapter, reads with poly-N sequences, and low quality reads from the raw data. Additionally, the Q20, Q30, GC content, and sequence duplication level were calculated for the clean reads, which were used in all downstream analyses48.
Functional annotation of genes
Gene functions were annotated based on the GO49 and KEGG pathway databases50.
Analysis of differentially expressed genes
Gene expression levels were estimated using the RSEM program51. Clean reads were mapped based on the assembled transcriptome, and the read count for each gene was obtained from the mapping results. Genes differentially expressed between two groups were analyzed using the DESeq R package (1.10.1), which provides statistical algorithms for detecting DEGs using a model based on a negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach to control the false discovery rate52. Genes with an adjusted P value < 0.05 following the DESeq analysis were considered to be differentially expressed between groups52.
Analysis of GO and KEGG pathway enrichment of differentially expressed genes
The GO enrichment analysis of the DEGs was completed using the GOseq R packages based Wallenius’ non-central hypergeometric distribution53, which can adjust for gene length biases among the DEGs. We used KOBAS software to test the significance of the enrichment of DEGs in specific KEGG pathways54.
Validation by qRT-PCR analysis
Leaves harvested from three independent seedlings of variegated and green samples of both Huangguogan and Shiranuhi were used as three biological replicates. Quantitative real-time PCR (qRT-PCR) analysis using the Bio-Rad CFX Manager (Bio-Rad, USA) with SsoFastTM EvaGreen Supermix (Bio-Rad) was employed to verify the DEG expression results23. Primers (Table 2) for specific genes encoding photosynthesis DEGs were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and synthesized by Sangon Biotech44. All primers were amplified with no template control to make sure the amplicons were not primer dimers. Gene expression levels were normalized against the geometric mean of citrus reference gene, Actin (GenBank: XM 006480741.2) and calculated by 2−ΔΔCT method55.
References
Biswas, M. K., Chai, L., Amar, M. H., Zhang, X. & Deng, X. Comparative analysis of genetic diversity in Citrus germplasm collection using AFLP, SSAP, SAMPL and SSR markers. Scientia Horticulturae 129, 798–803, https://doi.org/10.1016/j.scienta.2011.06.015 (2011).
Xiong, B. et al. Exogenous spermidine alleviates fruit granulation in a Citrus cultivar (Huangguogan) through the antioxidant pathway. Acta Physiologiae Plantarum 4, 98, https://doi.org/10.1007/s11738-017-2397-6 (2017).
Katinakis, P. Spatio-temporal changes in endogenous abscisic acid contents during etiolated growth and photomorphogenesis in tomato seedlings. Biotech Histochem 10, e1039213, https://doi.org/10.3109/10520295.2015.1020875 (2015).
Yuan, M. et al. Light Regulates Transcription of Chlorophyll Biosynthetic Genes During Chloroplast Biogenesis. Critical Reviews in Plant Sciences, 1–20, https://doi.org/10.1080/07352689.2017.1327764 (2017).
Mao, T. Light regulation of mitochondrial alternative oxidase pathway during greening of etiolated wheat seedlings. Plant physiology 174, 75–84, https://doi.org/10.1104/pp.15.00609 (2015).
Warpeha, K. M. The basal level ethylene response is important to the wall and endomembrane structure in the hypocotyl cells of etiolated Arabidopsis seedlings. Methods in molecular biology 54, 434–455, https://doi.org/10.1007/978-1-62703-532-3_10 (2012).
Hedtke, B. et al. Deficiency in riboflavin biosynthesis affects tetrapyrrole biosynthesis in etiolated Arabidopsis tissue. Cell research 78, 77–93, https://doi.org/10.1038/cr.2012.29 (2012).
Deng, X. W. Signaling role of phospholipid hydroperoxide glutathione peroxidase (PHGPX) accompanying sensing of NaCl stress in etiolated sunflower seedling cotyledons. Proceedings of the National Academy of Sciences of the United States of America 9, e977746, https://doi.org/10.1073/pnas.1402491111 (2014).
Quan, S. et al. Proteome Analysis of Peroxisomes from Etiolated Arabidopsis Seedlings Identifies a Peroxisomal Protease Involved in b-Oxidation and Development. Plant physiology, 1518–1538, https://doi.org/10.1104/pp.113.223453 (2013).
Ning, D. L. et al. Large-scale comparative phosphoprotein analysis of maize seedling leaves during greening. Planta 243, 501–517, https://doi.org/10.1007/s00425-015-2420-3 (2016).
Hung, C. Y. et al. Identification of a Mg-protoporphyrin IX monomethyl ester cyclase homologue, EaZIP, differentially expressed in variegated Epipremnum aureum ‘Golden Pothos’ is achieved through a unique method of comparative study using tissue regenerated plants. Journal of experimental botany 61, 1483–1493, https://doi.org/10.1093/jxb/erq020 (2010).
Kato, Y., Miura, E., Matsushima, R. & Sakamoto, W. White leaf sectors in yellow variegated2 are formed by viable cells with undifferentiated plastids. Plant physiology 144, 952–960, https://doi.org/10.1104/pp.107.099002 (2007).
Li, Q. et al. Proteomic analysis of young leaves at three developmental stages in an albino tea cultivar. Proteome science 9, 44, https://doi.org/10.1186/1477-5956-9-44 (2011).
Jensen, P. E. et al. Structural genes for Mg-chelatase subunits in barley: Xantha-f, -g and -h. Molecular & general genetics: MGG 250, 383–394 (1996).
Zhang, H. et al. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant molecular biology 62, 325–337, https://doi.org/10.1007/s11103-006-9024-z (2006).
Wu, Z. et al. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant physiology 145, 29–40, https://doi.org/10.1104/pp.107.100321 (2007).
Zhang, H. et al. Transcript profile analysis reveals important roles of jasmonic acid signalling pathway in the response of sweet potato to salt stress. Scientific reports 7, 40819, https://doi.org/10.1038/srep40819 (2017).
Qiao, Q. et al. Transcriptome sequencing of Crucihimalaya himalaica (Brassicaceae) reveals how Arabidopsis close relative adapt to the Qinghai-Tibet Plateau. Scientific reports 6, 21729, https://doi.org/10.1038/srep21729 (2016).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature protocols 8, 1494–1512, https://doi.org/10.1038/nprot.2013.084 (2013).
Usadel, B. et al. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant, cell & environment 32, 1633–1651, https://doi.org/10.1111/j.1365-3040.2009.02040.x (2009).
Ding, Z. et al. Identification of Photosynthesis-Associated C4 Candidate Genes through Comparative Leaf Gradient Transcriptome in Multiple Lineages of C3 and C4 Species. PloS one 10, e0140629, https://doi.org/10.1371/journal.pone.0140629 (2015).
Hansen, B. O., Vaid, N., Musialak-Lange, M., Janowski, M. & Mutwil, M. Elucidating gene function and function evolution through comparison of co-expression networks of plants. Frontiers in plant science 5, 394, https://doi.org/10.3389/fpls.2014.00394 (2014).
Xiong, B. et al. Transcriptome Analyses of Two Citrus Cultivars (Shiranuhi and Huangguogan) in Seedling Etiolation. Scientific reports 7, 46245, https://doi.org/10.1038/srep46245 (2017).
Storey, J. D. & Tibshirani, R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods in molecular biology 224, 149–157, https://doi.org/10.1385/1-59259-364-X:149 (2003).
Liu, Y. et al. Uniconazole-induced starch accumulation in the bioenergy crop duckweed (Landoltia punctata) I: transcriptome analysis of the effects of uniconazole on chlorophyll and endogenous hormone biosynthesis. Biotechnol Biofuels 8, 57, https://doi.org/10.1186/s13068-015-0246-7 (2015).
Nagata, N., Tanaka, R., Satoh, S. & Tanaka, A. Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. The Plant cell 17, 233–240, https://doi.org/10.1105/tpc.104.027276 (2005).
Gibson, L. C. & Hunter, C. N. The bacteriochlorophyll biosynthesis gene, bchM, of Rhodobacter sphaeroides encodes S-adenosyl-L-methionine: Mg protoporphyrin IX methyltransferase. FEBS letters 352, 127–130, https://doi.org/10.1016/0014-5793(94)00934-1 (1994).
Meguro, M., Ito, H., Takabayashi, A., Tanaka, R. & Tanaka, A. Identification of the 7-hydroxymethyl chlorophyll a reductase of the chlorophyll cycle in Arabidopsis. The Plant cell 23, 3442–3453, https://doi.org/10.1105/tpc.111.089714 (2011).
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic acids research 47, D590–D595, https://doi.org/10.1093/nar/gky962 (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research 28, 27–30, https://doi.org/10.1093/nar/28.1.27 (2000).
Song, L. L., Zhao, H. Q., Zhu, X. Q., Dong, G. X. & Xie, R. Effects of high temperature stress on photosynthesis and chlorophyll fluorescence of rice. Journal of Anhui Agricultural Sciences 39, 13348–13353 (2011).
Sicher, R. C. & Bunce, J. A. Adjustments of net photosynthesis in Solanum tuberosum in response to reciprocal changes in ambient and elevated growth CO2 partial pressures. Physiologia plantarum 112, 55–61 (2001).
Gao, M., Qi, Y., Song, W. & Xu, H. Effects of di-n-butyl phthalate and di (2-ethylhexyl) phthalate on the growth, photosynthesis, and chlorophyll fluorescence of wheat seedlings. Chemosphere 151, 76–83, https://doi.org/10.1016/j.chemosphere.2016.02.061 (2016).
Matsumoto, F. et al. Gene expression profiling of the tetrapyrrole metabolic pathway in Arabidopsis with a mini-array system. Plant physiology 135, 2379–2391, https://doi.org/10.1104/pp.104.042408 (2004).
Alawady, A. E. & Grimm, B. Tobacco Mg protoporphyrin IX methyltransferase is involved in inverse activation of Mg porphyrin and protoheme synthesis. The Plant journal: for cell and molecular biology 41, 282–290, https://doi.org/10.1111/j.1365-313X.2004.02291.x (2005).
Yang, Y. T., Yu, Y. L., Yang, G. D., Zhang, J. D. & Zheng, C. C. Tissue-specific expression of the PNZIP promoter is mediated by combinatorial interaction of different cis-elements and a novel transcriptional factor. Nucleic acids research 37, 2630–2644, https://doi.org/10.1093/nar/gkp126 (2009).
Bartsch, S. et al. Three thioredoxin targets in the inner envelope membrane of chloroplasts function in protein import and chlorophyll metabolism. Proceedings of the National Academy of Sciences of the United States of America 105, 4933–4938, https://doi.org/10.1073/pnas.0800378105 (2008).
McCormac, A. C. & Terry, M. J. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. The Plant journal: for cell and molecular biology 40, 672–685, https://doi.org/10.1111/j.1365-313X.2004.02243.x (2004).
Russell, A. W. et al. Photosystem II Regulation and Dynamics of the Chloroplast D1 Protein in Arabidopsis Leaves during Photosynthesis and Photoinhibition. Plant physiology 107, 943–952 (1995).
Nap, J. P. et al. Activity of the promoter of the Lhca3.St.1 gene, encoding the potato apoprotein 2 of the light-harvesting complex of Photosystem I, in transgenic potato and tobacco plants. Plant molecular biology 23, 605–612 (1993).
Berry, J. O., Breiding, D. E. & Klessig, D. F. Light-mediated control of translational initiation of ribulose-1, 5-bisphosphate carboxylase in amaranth cotyledons. The Plant cell 2, 795–803, https://doi.org/10.1105/tpc.2.8.795 (1990).
Zhao, X. X., Huang, L. K., Zhang, X. Q., Li, Z. & Peng, Y. Effects of heat acclimation on photosynthesis, antioxidant enzyme activities, and gene expression in orchardgrass under heat stress. Molecules 19, 13564–13576, https://doi.org/10.3390/molecules190913564 (2014).
Molnarova, M. & Fargasova, A. Se(IV) phytotoxicity for monocotyledonae cereals (Hordeum vulgare L., Triticum aestivum L.) and dicotyledonae crops (Sinapis alba L., Brassica napus L.). Journal of hazardous materials 172, 854–861, https://doi.org/10.1016/j.jhazmat.2009.07.096 (2009).
Xiong, B. et al. Variability in CitXET expression and XET activity in Citrus cultivar Huangguogan seedlings with differed degrees of etiolation. PloS one 12, e0178973, https://doi.org/10.1371/journal.pone.0178973 (2017).
Jia, D. et al. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt). Scientific reports 8, 2197, https://doi.org/10.1038/s41598-018-20181-y (2018).
Yu, S., Wang, G., Liao, J., Tang, M. & Sun, W. Transcriptome Profile Analysis of Mechanisms of Black and White Plumage Determination in Black-Bone Chicken. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology 46, 2373–2384, https://doi.org/10.1159/000489644 (2018).
Lai, D., Jin, X., Wang, H., Yuan, M. & Xu, H. Gene expression profile change and growth inhibition in Drosophila larvae treated with azadirachtin. Journal of biotechnology 185, 51–56, https://doi.org/10.1016/j.jbiotec.2014.06.014 (2014).
He, B. et al. Transcriptome analysis of Ginkgo biloba kernels. Frontiers in plant science 6, 819, https://doi.org/10.3389/fpls.2015.00819 (2015).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics 25, 25–29, https://doi.org/10.1038/75556 (2000).
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y. & Hattori, M. The KEGG resource for deciphering the genome. Nucleic acids research 32, D277–280, https://doi.org/10.1093/nar/gkh063 (2004).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 12, 323, https://doi.org/10.1186/1471-2105-12-323 (2011).
Yu, C. et al. Soil fungal community comparison of different mulberry genotypes and the relationship with mulberry fruit sclerotiniosis. Scientific reports 6, 28365, https://doi.org/10.1038/srep28365 (2016).
Young, M. D., Wakefield, M. J., Smyth, G. K. & Oshlack, A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome biology 11, R14, https://doi.org/10.1186/gb-2010-11-2-r14 (2010).
Mao, X., Cai, T., Olyarchuk, J. G. & Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793, https://doi.org/10.1093/bioinformatics/bti430 (2005).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408, https://doi.org/10.1006/meth.2001.1262 (2001).
Acknowledgements
This research was funded by grants: 2019NZZJ0013 (to B.X.) and 2016NZ0056 (to Z.-H.W.) from The Science and Technology Department of Sichuan Province, China. We thank Dr. William Yajima, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text. We also acknowledge the copyright permission of KEGG for the using of map00195 (http://www.kegg.jp/kegg/kegg1.html).
Author information
Authors and Affiliations
Contributions
B.X., X.Q. and Z.-H.W. conceived and designed the experiments. B.X., X.Q. and S.-J.H. carried out the experiments. X.-J.W., X.Z.and J.Z. analyzed the data. T.-T.D., T.W. and S.-C.L. contributed to samples collection. B.X. and X.Q. wrote the paper. Z.-H.W. and G.-C.S. revised the manuscript. All authors approved the final revision to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xiong, B., Qiu, X., Huang, S. et al. Physiological and transcriptome analyses of photosynthesis and chlorophyll metabolism in variegated Citrus (Shiranuhi and Huangguogan) seedlings. Sci Rep 9, 15670 (2019). https://doi.org/10.1038/s41598-019-52276-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-52276-5
This article is cited by
-
Transcriptomic analysis reveals biosynthesis genes and transcription factors related to leaf anthocyanin biosynthesis in Aglaonema commutatum
BMC Genomics (2023)
-
Chimeric deletion mutation of rpoC2 underlies the leaf-patterning of Clivia miniata var. variegata
Plant Cell Reports (2023)
-
Comparative transcriptomics reveals key genes contributing to the differences in drought tolerance among three cultivars of foxtail millet (Setaria italica)
Plant Growth Regulation (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.