Abstract
Using Sprague-Dawley rats (350–450 g; n = 61) and the recently updated Walker-Mason rat scald burn model, we demonstrated that Pseudomonas aeruginosa readily formed biofilms within full-thickness burn wounds. Following the burn, wounds were surface-inoculated with P. aeruginosa in phosphate-buffered saline (PBS), while sterile PBS was used for controls. On post-burn days 1, 3, 7, and 11, animals were euthanized and samples collected for quantitative bacteriology, bacterial gene expression, complete blood cell counts, histology, and myeloperoxidase activity. Robust biofilm infections developed in the full-thickness burn wounds inoculated with 1 × 104 CFU of P. aeruginosa. Both histology and scanning electron microscopy showed the pathogen throughout the histologic cross-sections of burned skin. Quantigene analysis revealed significant upregulation of alginate and pellicle biofilm matrix genes of P. aeruginosa within the burn eschar. Additionally, expression of P. aeruginosa proteases and siderophores increased significantly in the burn wound environment. Interestingly, the host’s neutrophil response to the pathogen was not elevated in either the eschar or circulating blood when compared to the control burn. This new full-thickness burn biofilm infection model will be used to test new anti-biofilm therapies that may be deployed with soldiers in combat for immediate use at the site of burn injury on the battlefield.
Similar content being viewed by others
Introduction
One of the greatest challenges facing the burn clinic is the complication of bacterial infection within the burn wound that may lead to more severe disease states, including sepsis. Scarcity of new antimicrobials, specifically anti-biofilm agents, further escalates the challenge posed by resistant microorganisms in treating burn wound infections. Several different animal models that seek to recapitulate the hallmarks of clinical burn wound infection are used to assist the development of novel antimicrobial therapies. The three most common burn wound infection in vivo models include pigs, mice, and rats1,2,3,4,5. Porcine models are often the best substitute for human skin, but their use for screening vast librarys of antimicrobials are hampered by high costs and ethical implications3. Mice offer the greatest range of benefits for characterizing aspects of the host’s response to burn infection, based on genetic mutants, but are often limited to acute (less than 48 hours post inoculation) burn wound infection before rapid onset of sepsis and death, potentially due to the requirement for injection of the high bacterial inoculums beneath the burn eschar to induce the burn infection6,7,8. Others have also included formulation of the pathogen in a biopolymer matrix to drive the infection towards a more chronic biofilm-like state9,10. However, these inoculation and exposure routes do not recapitulate what occurs in burn clinics and fail to mimic the natural development of infection in full-thickness burn injuries. Rats have played a significant role in the development and transition of antimicrobial burn wound treatments to clinics since the 1960’s, when the original Walker-Mason rat scald burn and surface infection model was described11,12,13,14,15,16.
The Walker-Mason rat scald burn model displays the classical features of invasive burn wound infection, typically observed with Pseudomonas aeruginosa using a natural surface route of infection, albeit with high inoculum levels of the pathogen, after induction of the burn injury. Importantly, studies using this model directly led to the development and clinical adoption of the ‘gold standard’ antimicrobial treatments for severe burn wounds, namely Sulfamylon® (mafenide acetate) and Silvadene® (silver sulfadiazine)12,17,18,19. These treatments greatly reduced the overall lethality of septic burn wound infection by P. aeruginosa in clinics20. While these antimicrobial therapies are proven effective in preventing infection, the immense number of daily person-hours and ease of application limits their utility outside a tier IV or V military treatment facility, including regional referral hospitals and hospitals within the United States18,21. Coupled with the difficulty in treatment deployability to forward operating theatres, any delay in application of the antimicrobial therapy to the burn injury may result in its decreased effectiveness, potentially due to the uninhibited development of bacterial biofilms within the burn eschar12.
We recently updated the Walker-Mason rat scald burn model in regards to skin preparation, pain relief, resuscitation regimen, and scald times for both deep partial- and full-thickness burn wound formation22. Our previous work showed that, within three days post-burn and surface inoculation with as little as 1 × 103 CFU, the opportunistic pathogen P. aeruginosa, when left unimpeded, forms biofilms within deep partial-thickness burn wounds. Development of biofilm within the eschar was characterized by significant upregulation of genes encoding alginate (alg8 and algE) and the iron binding siderophore pyoverdine (pvdS)23,24,25,26,27,28,29. These molecules contribute to the structure of the biofilm matrix as well as liberates bound iron from host tissues, respectively25,26. In this report, we expand our findings to include the infection kinetics and inoculum required of P. aeruginosa and its biofilm formation in full-thickness burn wounds. Similar to partial-thickness burn wounds, P. aeruginosa developed robust infections within the burn eschar, but the host’s neutrophil response to the pathogen seemed reduced compared to the partial-thickness burn infection, reported previously. Compared to partial-thickness burns, the invasiveness of the pathogen, leading toward sepsis, seemed heightened in the full-thickness burn injury by recovery of the pathogen in the blood in as little as 3 days post burn and inoculation most likely caused by greater burn wound tissue necrosis. Importantly, several key genes related to biofilm formation by P. aeruginosa were significantly upregulated in the tissue indicating development of mature biofilm in the full-thickness burn wound tissue.
Results
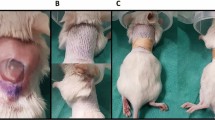
We characterized the kinetics of P. aeruginosa infection and biofilm formation in full-thickness scald burns using the modified Walker-Mason burn model in Sprague-Dawley rats. Figure 1 shows representative gross appearance of the burn wound with and without P. aeruginosa infection over the course of the study. Exudate became apparent by POD 3 in the wounds inoculated with P. aeruginosa. By POD 7 and 11, areas of tissue necrosis within the burn wound could be easily observed in both inoculum groups compared to the controls. The un-inoculated wounds did show contamination of the eschar based on the discoloration of the burned area. Given the severity of the wound infection, an expected decline in body weight was observed in both inoculum groups, but generally leveled off or began to recover during the study with the extra administrations of pain medication and Lactated Ringers on POD’s 3 and 7. The body weights of the animals over the course of the infection are shown in Supplementary Fig. 1.
Quantitative bacteriology
Viable CFU counts isolated from the tissue biopsies are shown in Fig. 2. A similar increase in bacterial number was obtained on P. aeruginosa isolation agar (Fig. 2A) and standard blood agar (Fig. 2B) as seen previously in partial-thickness burn wounds infected with P. aeruginosa22. By POD 7, a plateau in number of microorganisms, P. aeruginosa or native flora, per gram of wound tissue was observed. Large variability in the 1 × 103 CFU/wound inoculum was present in the counts obtained from P. aeruginosa isolation agar due to lack of P. aeruginosa recovery in the second run of the infection study. The results of the viable counts were confirmed via qPCR for bacterial DNA isolated from the homogenate of the same biopsy punches, shown in Fig. 3A,B. PCR analysis showed near perfect mirroring of the viable CFU counts. As a result of the variability within the 1 × 103 inoculum, the P. aeruginosa percentage of total bacterial DNA recovered from the wound sample was greatly reduced compared to the 1 × 104 inoculum, shown in Fig. 3C.
Maximum bacterial load of full-thickness burn wounds was achieved within 7 days. The viable counts from P. aeruginosa isolation agar (A) and non-selective agar (B) increase with time following the burn. P. aeruginosa was not cultured from un-inoculated control wounds. The 1 × 103 inoculum showed pronounced variability in the P. aeruginosa counts due to lack of recovery during the second experimental run. The 1 × 104 inoculum exhibited superior reproducibility, in terms of bacterial recovery, between all three experimental runs. Regardless of inoculation amount, by POD 11, all groups on the non-selective agar medium obtained a plateau of ~1 × 109 CFU/g wound tissue of either P. aeruginosa or native flora.
qPCR confirmed the P. aeruginosa and total bacterial loads within the burn eschar over 11 days following burn injury. The results of the qPCR analysis mirrored the data obtained in the viable counts for P. aeruginosa (A) and total bacterial cells (B). As a percentage of the total bacterial load (C), within three days post-burn and infection P. aeruginosa dominated the burn wounds inoculated with 1 × 104 CFU/wound. As expected, wounds inoculated with 1 × 103 CFU/wound showed greater variability of P. aeruginosa percentage of bacterial load due to its lack of recovery in the second experimental run.
Table 1 documents the results of the SignalTM Blood Culture System from the second and third runs of the study. Unfortunately, the Blood Culture System was not employed during the first experimental run, therefore blood culture results are not available from those animals. No animal showed positive results on POD 1. One of four animals in both inoculum groups showed positive blood cultures for P. aeruginosa on POD 3. By POD 7, 25% of 1 × 103 CFU inoculum group was positive for P. aeruginosa in the blood stream, while 50% of the 1 × 104 CFU inoculum group showed the pathogen in the blood. By POD 11, half of the animals in the 1 × 103 CFU inoculum group showed P. aeruginosa and Staphylococcus sciui ss sciuri growth in the blood, while all animals in the 1 × 104 CFU inoculum group were positive for P. aeruginosa or Staphylococcus xylosus in the blood. These data demonstrate that P. aeruginosa burn wound infection may lead to bacteremia by P. aeruginosa or other native flora30,31 [Sanjar et al. “Identification of Metagenomics Structure and Function Associated with Temporal Changes in Rat (Rattus norvegicus) Skin Microbiome during Health and Cutaneous Burn” in press] common to the rat skin that transgress to the blood stream within a week of the burn, if left untreated. Importantly, animals that received a burn but were not inoculated with P. aeruginosa did not show the same invasive burn wound infection and presence of any bacterial cells, P. aeruginosa or native microflora, in the blood stream.
Histology
We evaluated histologic cross-sections of the burn wound inoculated with P. aeruginosa by scanning electron microscopy (SEM). Figure 4A shows both 1,000× and 10,000× magnification views obtained by SEM of a wound inoculated with 1 × 104 CFU of P. aeruginosa over the course of the study. On POD 1, few if any bacterial cells were observed, but large numbers were seen penetrating into the eschar by POD 3. Similar to our work in partial-thickness burns, the bacterial cells were often associated with the collagen fibers of the burned tissue. We also examined the appearance of the eschar surface with SEM on POD 11. Figure 4B shows areas of both exposed bacterial cells and smoothed matrix like material that may be a combination of host proteins/tissues and bacterial exopolysaccharides of a wound inoculated with 1 × 104 CFU of P. aeruginosa. Additionally, upon high magnification (10,000×), the layers of bacterial cells could be easily seen in the cracks on the surface of the eschar, potentially a product of the sample dehydration during the preparation for SEM imaging. Unlike the wounds infected with P. aeruginosa, SEM images of control burn wounds inoculated with PBS, Supplementary Fig. 2, do not show the presence of bacterial cells associated with the collagen fibers or within a matrix on the wound surface.
Scanning electron microscopy revealed the penetration of P. aeruginosa deep into the burn eschar via histologic cross-sections (A) of burn wounds inoculated with 1 × 104 CFU/wound as well as dense biofilm matrix on the burn surface on POD 11 of the same inoculum group (B). In the tissue cross-sections, P. aeruginosa (arrows) invaded completely through the dermis and were associated with collagen fibers. Abundant numbers of P. aeruginosa (arrows) located on the surface of the burn were enmeshed in a smooth matrix material, possibly derived from both bacterial and host polysaccharides. Scale bar of 1,000× images is 10 µm and 10,000× images is 2 µm.
Micrographs of histologic sections from 1 × 104 inoculated wounds on POD 7 stained with H&E and Giemsa or PNA-FISH are shown in Fig. 5. Micrographs of histologic sections from control un-inoculated wounds are shown in Supplementary Fig. 3 and an overview of the tissue sections are displayed in Supplementary Fig. 4. High magnification images taken with the 63× objective of three areas in the tissue section include the epidermis, the middle of the dermis, and of the deep dermis near the panniculus carnosus. The combination of the brightfield stains allows for identification of the tissue structure while also highlighting the penetration of the pathogen throughout all levels of the burned eschar. PNA-FISH, specific for P. aeruginosa, confirmed the identity of the pathogen within the tissue sections. Respective micrographs of the un-inoculated wounds on POD 7 after the burn are shown in Supplementary Fig. 3. The un-inoculated wounds did show the presence of the native flora, mostly gram-positive cocci, within the histological sections stained with H&E and Giemsa, but lacked the presence of the P. aeruginosa and microcolonies observed in the inoculated wounds stained with PNA-FISH.
Representative micrographs of burn wound tissue cross-sections inoculated with 1 × 104 CFU of P. aeruginosa cells reveal penetration of individual P. aeruginosa cells (arrows, stained blue in H&E/Giemsa and red in PNA-FISH) at all levels (Epidermis, Mid-Dermis, and Deep-Dermis) of the burned tissue by day 7 post-burn and inoculation. Bacterial aggregates (arrowheads) were observed at the epidermis, middle of the dermis, and in the deep dermis near the panniculus carnosus. Scale bar is 40 µm.
Biofilm gene expression
Expression patterns of important biofilm associated genes of P. aeruginosa over the 11-day experiment are shown in Fig. 6 with results shown as a fold change of the planktonic inoculum. Alginate genes (algD, alg8, and algE) displayed in Fig. 6A–C show roughly 5–10 fold increases in expression levels as compared to the inoculum. Plateaus in algD and algE gene expression were reached by POD 7 after the burn. Alg8 showed higher expression levels in the 1 × 103 CFU/wound inoculated group as compared to the 1 × 104 CFU/wound group, which were uniformly elevated ~10 fold higher than the inoculum throughout the experiment. Similar expression increases were observed in the pelB, pelC, and pelD (pellicle biofilm) genes over the course of the study as depicted in Fig. 6D–F. Roughly, 5–15 fold increases in pellicle biofilm gene expression were detected in the burn wound compared to the inoculum, although pelC and pelD showed increased variability in the 1 × 103 inoculum. We measured the expression of three classical P. aeruginosa virulence genes (protease genes lasA, lasB, and iron starvation sigma factor gene pvdS) in the burn wound tissue, shown in Fig. 6G–I. Increased expression, between 10–30 fold over the inoculum, were measured over the course of the study for each virulence gene. Conversely, biofilm polysaccharide genes pslA and pslC did not show similar increases in expression level upon in vivo inoculation as seen in Fig. 6J,K, and may be reduced compared to the planktonic inoculum.
Increased expression of important P. aeruginosa biofilm genes detected in full-thickness burn tissue compared to the planktonic inoculum. Quantigene analysis showed increased expression of genes controlling production of alginate, algD (A), alg8 (B), and algE (C); pel polysaccharide, pelB (D), pelC (E), and pelD (F); and virulence genes, lasA (G), lasB (H), and pvdS (I) in the full-thickness burn wound tissue compared to the planktonic inoculum. Interestingly, expression of psl polysaccharide genes pslA (J) and pslB (K) did not increase in the burn wound environment. (*p < 0.05; **p < 0.01, ***p < 0.001 in comparison with the planktonic inoculum).
Host neutrophil response
The local tissue and systemic neutrophil response to the burn and P. aeruginosa infection are shown in Fig. 7. MPO activity confined to the burn wound, shown in Fig. 7A, was the same between all inoculation groups on each day. Generally, the MPO activity increased with time after the burn, but addition of P. aeruginosa did not further elevate Neutrophil MPO activity in the local tissue, regardless of inoculation level. Similar results were observed on a systemic level in the Neutrophil counts obtained from cardiac puncture as seen in Fig. 7B with only the 1 × 104 CFU/wound inoculum showing increased circulating Neutrophils as compared to the un-inoculated burn wounds on POD 11.
P. aeruginosa infection did not exacerbate the neutrophil response to full-thickness burn injury. General trends of increase MPO activity (A) were observed in full-thickness burn tissue, but were unaffected by concomitant P. aeruginosa infection. Slightly increased levels of circulating neutrophils (B) were measured following full-thickness burn injury but were not significantly increased with the presence P. aeruginosa.
Discussion
We previously updated the Walker-Mason rat scald burn model to document the growth of P. aeruginosa biofilms in deep partial-thickness burn wounds22. This current study builds on that foundation, extending our P. aeruginosa infected animal models to include full-thickness burn injuries. In our burn time optimization study, reported previously, we identified that the time required to achieve a uniform full-thickness burn in Sprague-Dawley rats was 6 s at 99 °C. To develop biofilm infections in the partial-thickness burn model, we inoculated the wounds with three different levels of P. aeruginosa, namely 1 × 103, 1 × 104, 1 × 105 CFU/wound. Given the concerns on animal survival with large (1 × 106 CFU/wound) P. aeruginosa inoculums in full-thickness burn injuries shown in earlier studies11,12,32, we limited the inoculum to only 1 × 103 and 1 × 104 CFU/wound. The reduced pathogen numbers avoided the lethality associated with greater bacterial inoculums as documented in previous literature11,12,13,32. We also included additional administration of proactive pain relief (Buprenorphine SR LAB) and lactated Ringers resuscitation to all animals remaining in the study beyond PODs 3 and 7. These additional treatments were provided to every animal in those groups in order to maintain uniformity between the different study groups.
Similar to partial-thickness burn wounds, we observed robust infections develop in all the full-thickness scald burns, plateauing above 1 × 108 CFU/g of wound tissue. The wounds inoculated with sterile PBS showed the growth of the native microflora in the burn eschar without any culturable P. aeruginosa over the 11 days of the study. Compared to partial-thickness burn wounds infected with P. aeruginosa, less tissue separation and sloughing was observed in the full-thickness burn wounds on PODs 7 and 11. However, the full-thickness burns infected with P. aeruginosa showed a greater degree of tissue necrosis as seen in the gross wound images in Fig. 1. Interestingly, the wounds inoculated with P. aeruginosa showed greater variability in recoverable CFU’s of P. aeruginosa compared to our previous work with partial-thickness burns. In the second experimental run, the bacterium was notably absent on both P. aeruginosa isolation and standard blood agar plates of the 1 × 104 CFU inoculum on POD 1 and the 1 × 103 CFU inoculum on PODs 3, 7, and 11. This was also confirmed by qPCR for the bacterial DNA indicating that this was not a plating error. However, the bacterium was observed in histological sections of those samples as well as at low levels in end point PCR for the oprL gene from different biopsy punches from the same burn wound. Presumably, in that experimental run the native flora may have suppressed the growth or recovery of P. aeruginosa, because the first and third runs showed greater recovery of the pathogen from the biopsy punch samples of the 1 × 103 CFU/wound inoculum group. Therefore, sampling errors could be a contributing factor to the recovery of the pathogen in that study group. Conversely, the 1 × 104 CFU/wound inoculum showed better consistency between all three runs, suggesting an optimal infectious dose of P. aeruginosa to surface infect full-thickness burn wounds without the mortality observed in previous studies11,12. This inoculation method is important because it did not require injection of the pathogen under the burn eschar to establish the biofilm infection, which is commonly used in many murine and rodent burn infection studies6,7,9,10. Additionally, the inoculum was spread over the burn surface as planktonic cells and did not require the use of a biomaterial10. This model recapitulated the invasive P. aeruginosa burn wound infection using a much lower inoculum of the pathogen than previously used, potentially mimicking a more clinically relevant burn infection model than others described11,12.
We utilized a new assay that directly measures mRNA to determine expressional changes of important biofilm and virulence associated genes of P. aeruginosa in the context of full-thickness burn wound infection. The QuantiGene technology selectively binds mRNA and amplifies the signal of the capture beads specific for the gene of interest. This method avoids the typical challenges associated with reverse transcription of mRNA into cDNA used in qRT-PCR, but instead directly binds to the target mRNA produced by the bacterial cells. The results were expressed as fold change over the planktonic inoculum. The genes involved in alginate biosynthesis (algD and alg8) and export (algE) increased throughout the infection. This indicated that P. aeruginosa produced and exported the exopolysaccharide alginate, an important constituent of its mature biofilm matrix. Similar increases occurred in genes associated with biofilm matrix component pel (pelB, pelC, and pelD). However, little to no increase in psl exopolysaccharide (pslA and pslC) production occurred in the infection. This may be due to the competence this clinical P. aeruginosa strain has for producing psl. Similar differences in pel and psl expression have been reported in laboratory P. aeruginosa strains PAO1 and PA1424,33,34,35,36. These two P. aeruginosa strains generally express different biofilm matrix components, namely psl (PAO1) and pel (PA14). The strain used in this study may possibly be more pel dominant or potentially deficient for producing psl. We previously characterized the the genome sequence of this clinically isolated pathogen37, but work is currently ongoing to further understand the in vitro characteristics of this pathogen namely its biofilm formation, motility, and antibiotic resistance genes compared to several other clinical and laboratory isolates of P. aeruginosa (manuscript in preparation). Lastly, expression of three virulence genes (lasA, lasB, and pvdS) all showed increases upon in vivo infection, indicating adaptation to the wound environment by increasing production of siderophores to liberate iron from host tissues and elastase to degrade the host tissue matrix38,39,40,41. Scavenging bound iron from the blood and tissues within the burn wound is an important step for P. aeruginosa biofilm formation42,43. Taken together the increased expression of biofilm exopolysaccharides along with factors that degrade host tissues and liberate important nutrients indicates active biofilm development by P. aeruginosa in full-thickness burn wounds.
Initial characterization of the host response to the tissue damage resulting from the full-thickness burn and subsequent P. aeruginosa infection suggests marked differences from partial-thickness burns. Our work in deep partial-thickness burns showed both a local and systemic, inoculum-dependent, neutrophil response to the P. aeruginosa infection. Interestingly, in the full-thickness burn, a similar elevation of neutrophil activity with P. aeruginosa infection, regardless of inoculum level, above the burn trauma itself was notably absent. A general trend of increased MPO activity was observed as time progressed in response to the burn, but not necessarily to the presence of P. aeruginosa in the burn eschar. Additionally, only on POD 11 was an increase in systemic neutrophil counts seen in only the 1 × 104 CFU/wound inoculum group as compared to all the other inoculation groups. The increased neutrophil counts were accompanied with a large standard deviation, because the two samples from the final experimental run of the 1 × 104 CFU/wound inoculum clotted before they could be analyzed, while the first run showed lower counts (~10 × 103 neutrophils/µl) and the second run had higher counts (~34 × 103 neutrophils/µl). Comparatively, in the partial-thickness burn with P. aeruginosa infection, we previously reported a trend of increased systemic neutrophils by POD 7 and significant increases on POD 1122. The lack of neutrophil response to the subsequent infection of full-thickness burns with P. aeruginosa may be due to the lack of blood flow or circulation in the burn eschar. In the deep partial-thickness burn model, roughly the bottom third of the dermis remained vitalized, while the full-thickness burn dermis was devoid of blood circulation. In larger TBSA rat burn models, slow accumulation of neutrophils into full-thickness burned skin, due to limited number of blood vessel beneath the burned dermis, has previously been shown44. Similarly, sequestration of neutrophils into distal injury sites (polyurethane sponges or bacterial skin lesions) from the burn was reduced with concomitant presence of full-thickness burn injuries45,46. These intrinsic features of the burn wound, albiet in larger TBSA burns, may have reduced the host’s ability to interact with P. aeruginosa or native microflora in the burn injury, thereby shielding the invading microbes from the innate immune response. Further, in-depth analysis (local and systemic cytokine expression, histologic scoring of infected verus un-inoculated tissue sections, circulating damage/pathogen associated molecular patterns, and systemic immune cell counts) of the host response to both partial- and full-thickness burns in the context of P. aeruginosa biofilm burn wound infection will be reported in a future manuscript.
In this report, we detail the course of P. aeruginosa infection in a 10% TBSA full-thickness scald burn with Sprague-Daweley rats. Robust infections developed using relatively low inoculums of 1 × 104 P. aeruginosa cells spread over the surface of the burn wound instead of injecting the inoculum beneath the burn eschar. The bacterial infection resulted in the natural development of P. aeruginosa biofilms in the burn eschar, coupled with systemic infection by 11 days post burn and inoculation. This work further enhances our portfolio of rat scald burn biofilm infection models for testing the effectiveness of novel and easily deployable anti-biofilm treatments in the context of both severe partial- and full-thickness burn wounds that afflict combat soldiers on the battlefield.
Materials and Methods
Animal ethics statement
Research was conducted in compliance with the Animal Welfare Act, the implementing Animal Welfare Regulations, and the principles of the Guide for the Care and Use of Laboratory Animals, National Research Council. The United States Army Institute of Surgical Research (USAISR) Institutional Animal Care and Use Committee approved all research conducted in this study (Animal Protocol A-16–047) on 15 September 2016. The facility where this research was conducted is fully accredited by AAALAC International.
Full-thickness burn and infection
Full-thickness burns were made on the dorsum of 61 male Sprague-Dawley rats using the method we previously described22. The burn area averaged ~10% of the total body surface area based on a body weight range between 350–450 g in Meeh’s formula, \(A=k{W}^{\frac{2}{3}}\), where A is total body surface area (cm2), k is Meeh’s constant (9.46), and W is body weight (g)47. In brief, the animals were acclimated to the facility for 2 weeks prior to the burn. The day before the burn, the dorsal surface of anesthetized (Forane, Baxter Healthcare Corporation, Deerfield, IL) rats was shaved and depilated (Nair, Church & Dwight Co, Ewing, NJ). Additionally, 1.2 mg/kg of Buprenorphine SR LAB (Zoopharm Pharmacy) was subcutaneously administered for proactive pain management. Post-shaving, the rats were housed individually until the end of the experiment.
Immediately prior to the burn, rats were anesthetized with 2.5% isoflurane (Forane) for 15 minutes in an induction chamber followed by application of eye lube (Artificial Tears Ointment, Akorn, Inc., Lake Forrest, IL). Pulse rate and blood oxygen levels (2500A Vet Pulse Oximeter, Nonin Medical, Inc., Plymouth, MN) were monitored throughout the procedure. The anesthetized rats were positioned into a custom burn template, which was lowered into 99 °C water bath for 6 seconds. We previously showed that this exposure time produced full-thickness burns in Sprague-Dawley rats22. Immediately post-burn, the burn area was blotted on damp (room temperature) paper towels to remove excess water and the first of four fluid resuscitations (warm 4 ml Lactated Ringers Solution (LRS) based on Parkland’s Formula) given via intraperitoneal injection. The remaining resuscitations were given within 36 hours following the burn. Burn wounds were imaged using a Nikon D90 with Nikkor AF-S lens (18–105 mm, A:3.5–5.6 G) following the burn and on the day of euthanasia.
Immediately after the burn and first resuscitation, P. aeruginosa strain 12-4-4(59) suspended in 1 × phosphate-buffered saline (PBS) was inoculated on the burn surface at 1 × 103 or 1 × 104 CFU/wound11,37. The bacterial cells were grown overnight at 37 °C in Trypticase Soy Broth (TSB; Becton, Dickerson and Company, Sparks, MD) and subcultured the day of the burn in fresh TSB to mid-logarithm growth phase. Prior to inoculation, the bacterial cells were centrifuged at 3,000 × g for 15 minutes at 4 °C and re-suspended in PBS to the correct cell density. A 100 µl P. aeruginosa suspension was spread over the burn area with a sterile pipette tip. Un-inoculated control wounds received 100 µl of sterile PBS. In total 61 animals comprised the study, with 12 serving as un-inoculated controls and 24 in each P. aeruginosa inoculum group. The study was repeated three times. Each experimental repeat consisted of one third of the animals in each group, distributed between four end points, post-operative day (POD) 1, 3, 7, and 11. One animal in the 1 × 104 CFU/wound inoculum required a replacement due to early removal from the study on POD 5.
To prevent disruption of the burn surface by the animals, the wounds were sealed with TegadermTM Film (3M Health Care, St. Paul, MN) using NOTAPE professional silicone bonding adhesive (Vapon, Inc. Fairfield, NJ). N-terface Wound Contact Layer (Winfield Laboratories, Inc. Richardson, TX) separated the burn surface and the TegadermTM Film. Rats were additionally placed into composite rat jackets, described previously for the duration of the study22. A ThermoCare Portable Animal Intensive Care Unit (Daisy Products LLC, Paso Robles, CA) set to 37 °C was used to recover the animals after the burn until the effects of the anesthesia wore off. Additional fluid resuscitation (LRS given intraperitoneally) and pain relief (Buprenorphine SR LAB given subcutaneously) beyond the initial 36 hours following the burn were provided on PODs 3 and 7 to rats remaining in the study beyond those time points. Daily evaluation of body weight and symptoms of pain/distress were made throughout the study.
End point procedures
For euthanasia, rats were anesthetized with 100 mg/kg Ketamine HCl (Zetamine, MWI Veterinary Supply Co. Boise, ID) and 10 mg/kg Xylazine (Akron Animal Health, Inc. Lake Forrest, IL) administered intraperitoneally. Isoflurane (4%) was given as needed during the terminal procedure. A cardiac puncture was used to obtain a blood sample for systemic neutrophil counts (EDTA Vacutainer 367841, Becton, Dickerson and Company, Sparks, MD) using an Abbott CELL-DYN® 3700 Blood Count Analyzer (Abbott Laboratories, Abbott Park, IL). Additionally, in the second and third runs of the study, 2.0 ml of blood was aseptically aliquoted into OxoidTM SignalTM Blood Culture System bottles (Thermo Fisher Scientific, Waltham, MA) to detect the presence of bacterial cells in the systemic circulation. Bacterial cells found in the systemic circulation were identified using a GEN III OmniLog® Combo System (Biolog, Inc. Hayward, CA). Fatal-Plus® (Vortech Pharmaceuticals, LTD. Dearborn, MI) was administered intra-cardiac and euthanasia confirmed by lack of cardiac movement, pulse, and breathing as defined by the AVMA Guidelines for the Euthanasia of Animals: 2013 Edition48.
Upon removal of the Tegaderm and N-terface dressings, the wounds were imaged and entire burn area excised from the dorsum. Three (cranial, middle, and caudal) separate histologic cross-sections of the burn area were fixed in 10% buffered formalin in PBS (Fisher Diagnostics, Kalamazoo, MI) for 48 hours and processed for routine embedding in paraffin wax. Biopsy punch (7 mm) samples of the burn area were isolated for quantitative bacteriology, scanning electron microscopy, myeloperoxidase (MPO) activity, and bacterial mRNA analysis. Internal organs (spleen, liver, kidney, and lungs) were also recovered. Tissue and organ samples, except those for quantitative bacteriology, were flash frozen in liquid Nitrogen and kept at −80 °C prior to analysis.
Quantification of bacterial load
The samples (biopsy punches) used to determine the bacterial load within the burn tissue were homogenized in 1.0 ml of PBS in MagNA Lyser Green Beads tubes (Roche Diagnostics GmbH, Mannheim, Germany) using a FastPrep®-24 Tissue Homogenizer (MP Biomedicals, LLC. Santa Ana, CA). The homogenized samples were plated in duplicate on Trypticase soy agar containing 5% sheep’s blood (Becton, Dickinson and Co. Sparks MD) and P. aeruginosa isolation agar (Hardy Diagnostics, Santa Maria, CA). Viable bacterial colonies were counted and plotted as log10 (CFU/g wound tissue) ± Standard Deviation.
Bacterial DNA was also isolated from the same biopsy punch samples for viable CFU quantification using PCR as previously described22. Fifty microliters from each of the four-biopsy sample tissue homogenates per animal were pooled and DNA isolated using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA). To quantify bacterial load in the burn eschar, primers and probes for outer membrane lipoprotein oprL of P. aeruginosa49 and universal primers and probes for the 16S rDNA of gram-negative bacterial cells were used50. Primer and probe sequences are listed in Supplemental Table 1. Standard curves were generated from mid-log growth phase cultures of P. aeruginosa. Genomic DNA concentration was measured using a Quant-iT ds DNA BR Assay Kit (Invitrogen, Carlsbad, CA) and confirmed using gel separation. All samples were run in triplicate on a StepOne Plus Real-Time PCR System (Applied Biosystems, Inc. Foster City, CA) using a total volume of 20 µl of TaqMan Gene Expression Master Mix (Applied Biosystems). Concentrations of forward and reverse primers and TaqMan MGB probe were set to 100 nM along with 5 µl of sample template. Target DNA was amplified using a standard qPCR protocol of a 5 minute 95 °C denaturation step, followed by 40 PCR amplification cycles (95 °C for 15 s and 60 °C for 60 s). The resulting data was analyzed using the StepOne software provided with the instrument. The genome copy number was calculated from the amount of genomic (P. aeruginosa or total) DNA and normalized by the weight of the tissue to provide CFU/g tissue.
Histology
Thin 5 µm sections of fixed rat burn eschar were stained with hematoxylin-eosin (H&E) or H&E with Giemsa to evaluate tissue damage and P. aeruginosa invasion into the tissue. Images were taken with both the 20× and 63× objectives of a Leica Aperio Versa 200 slide scanner (Leica Biosystems, Inc. Buffalo Grove, IL). Additionally, peptide nucleic acid (PNA)-fluorescent in situ hybridization (FISH) was used to confirm the presence of P. aeruginosa deep within the burned tissue section using a standardized method in our laboratory51. The E. coli/P. aeruginosa PNA FISH® Kit was used to visualize the penetration of P. aeruginosa into the burn eschar. Briefly, tissue sections were incubated with AvanDx fixative solution for 20 minutes at 55 °C. The PNA-FISH probe was incubated with each tissue section for 45 minutes allowing hybridization of the probe to the bacterial cells. The tissue sections were then washed with wash buffer, supplied with the kit, for 45 minutes. Tissue sections were mounted with the supplied mounting medium and edges of the coverslip sealed with nail polish. Specimens were imaged using the Texas Red filter (596/615 nm) and DAPI filter (358/461) with both 20× and 63× objectives of the Leica Aperio Versa 200 slide scanner.
Scanning electron microscopy
Both the surface and tissue cross-section of the burn eschar were evaluated by scanning electron microscopy (SEM). To visualize the surface of the wound, seven millimeter biopsy punch samples were fixed with 2.5% phosphate-buffered glutaraldehyde for at least 24 hours at 4 °C. Fixed specimens were processed through a graded series dehydration using cold ethanol/water (10%, 30%, 50%, 70%, 80%, 90%, 95%, 100%) and an incubation time of 10 minutes followed by critical point drying (EM CPD300, Leica Biosystems Inc. Buffalo Grove, IL). Immediately prior to examination, the wound biopsy punch samples were coated with carbon and gold/palladium (Leica ACE600 Coater). The histological cross-sections were deparaffinized with 100% xylene and air dried prior to coating with carbon and gold/palladium. Both sets of specimens (biopsy punches and tissue cross-sections) were visualized using a Sigma VP40 field emission scanning electron microscope (Carl Zeiss, Inc. Germany) in high vacuum mode at 2 kV.
QuantiGene plex gene expression assay for key P. aeruginosa biofilm and virulence genes
We used the QuantiGene Plex Assay (Assay ID M17102001, Affymetrix, Inc. Santa Clara, CA) to determine the expression level of key biofilm structural genes (algD, alg8, algE, pelB, pelC, pelD, pslA, and pslC) and virulence genes (lasA, lasB, and pvdS) using three distinct housekeeping genes (proC, fabD, and rpoD) following the manufacturer’s procedure for Frozen Tissue Homogenates. Briefly, samples were pulverized under liquid Nitrogen using a Bessman Tissue Pulverizer (Spectrum, Inc. Rancho Dominguez, CA) and lysed for 15 minutes at 65 °C with lysis mixture (cold TES Buffer with Lysozyme and Proteinase K Homogenization solution). The supernatants of the tissue homogenates were stored at −80 °C, after centrifuging at 4,000 rpm for 5 minutes, until analysis. The tissue homogenates were thawed and incubated at 37 °C for 30 minutes prior to mixing with the Working Bead Mix, containing Proteinase K and the Capture Beads, in a hybridization plate and incubated at 54 °C for 18 hours with gentle shaking at 600 rpm. After the overnight incubation, the sample was transferred to the Magnetic Separation Plate. Using the Hand-held Magnetic Plate Washer, the samples were washed 3× with Wash Buffer in between 1 hour incubations at 50 °C with Pre-Amplifier Solution, Amplifier Solution, and Label Probe Solution. After the Label Probe Solution, SAPE Working Reagent was added for 30 minutes at room temperature. After washing the plate 3× with SAPE Wash Buffer, 130 µl of SAPE Wash Buffer was added to each sample and mixed for 3 minutes at 800 rpm followed by immediate reading on a Bio-Plex 200 System with Bio-Plex ManagerTM Software Version 6.1 Build 727 (BioRAD Laboratories, Inc. Hercules, CA).
Quantification of myeloperoxidase activity in burn tissue
A Bessman Tissue Pulverizer was used to fracture biopsy punch samples under liquid Nitrogen. The samples were further homogenized as directed by the Fluoro MPO Myeloperoxidase Detection Kit (Cell Technology, Inc. Mountain View, CA). Briefly, an IKA T10 basic Ultra Turrax tissue homogenizer (IKA Works, Inc. Wilmington, NC) was used to process the samples in MPO homogenization buffer, provided in the kit. The samples were then centrifuged at 12,000 × g for 20 minutes at 4 °C and re-suspended in MPO solubilization buffer containing 0.5% Hexadecyltrimethylammonium (Sigma-Aldrich, Inc. St. Louis, MO). The re-suspended samples were homogenized again and sonicated using a Sonic Dismembrator Model 100 (Fisher Scientific, Kalamazoo, MI) for 30 s followed by two cycles of freezing/thawing before isolating the supernatant. Equal volumes of sample or MPO enzyme standard were mixed with MPO reaction cocktail and incubated in black opaque, clear bottom, microtiter plates for 30 minutes at room temperature. Fluorescence (530/590 nm) was measured using a BioTek Synergy HT with Gen5 Software (BioTek Instruments, Inc. Winooski, VT). MPO activity of each sample was calculated from the MPO enzyme standard curve and plotted as MPO activity in milliUnits (mU) ± Standard Deviaiton.
Statistical analysis
All data were plotted and analyzed using GraphPad Prism 7.03 (GraphPad Software, Inc. San Diego, CA). Comparisons were performed using two-way ANOVA with Tukey’s correction for multiple comparison and an α set to 0.05. Data was graphed as the mean ± Standard Deviation (SD).
Disclaimer
The opinions or assertions contained herein are the private views of the author and not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Dai, T. et al. Animal models of external traumatic wound infections. Virulence. 2(4), 296–315 (2011).
Abdullahi, A., Amini-Nik, S. & Jeschke, M. G. Animal models in burn research. Cell Mol Life Sci. 71(17), 3241–55 (2014).
Qu, M. & Nourbakhsh, M. Current experimental models of burns. Discov Med. 23(125), 95–103 (2017).
Guillory, A. N. et al. Cardiovascular Dysfunction Following Burn Injury: What We Have Learned from Rat and Mouse Models. Int J Mol Sci. 17(1) (2016).
Cornforth, D. M. et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci USA 115(22), E5125–e5134 (2018).
Rumbaugh, K. P. et al. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect Immun. 67(11), 5854–62 (1999).
Rumbaugh, K. P. et al. The effects of infection of thermal injury by Pseudomonas aeruginosa PAO1 on the murine cytokine response. Cytokine. 16(4), 160–8 (2001).
Turner, K. H. et al. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet. 10(7), e1004518 (2014).
Calum, H. et al. Thermal injury induces impaired function in polymorphonuclear neutrophil granulocytes and reduced control of burn wound infection. Clin Exp Immunol. 156(1), 102–10 (2009).
Trostrup, H. et al. Pseudomonas aeruginosa biofilm hampers murine central wound healing by suppression of vascular epithelial growth factor. Int Wound J. 15(1), 123–132 (2018).
Walker, H. L., Mason, A. D. Jr. & Raulston, G. L. Surface Infection with Pseudomonas Aeruginosa. Ann Surg. 160, 297–305 (1964).
Lindberg, R. B., Moncrief, J. A. & Mason, A. D. Jr. Control of experimental and clinical burn wounds sepsis by topical application of sulfamylon compounds. Ann N Y Acad Sci. 150(3), 950–60 (1968).
Yurt, R. W. et al. Increased susceptibility to infection related to extent of burn injury. Arch Surg. 119(2), 183–8 (1984).
Chu, C. S. et al. Topical silver treatment after escharectomy of infected full thickness burn wounds in rats. J Trauma. 58(5), 1040–6 (2005).
McManus, A. T., Moody, E. E. & Mason, A. D. Bacterial motility: a component in experimental Pseudomonas aeruginosa burn wound sepsis. Burns. 6(4), 235–239 (1980).
Rabin, E. R. et al. Fatal pseudomonas infection in burned patients. A clinical, bacteriologic and anatomic study. N Engl J Med. 265, 1225–31 (1961).
Chu, C. S. et al. Therapeutic effects of silver nylon dressings with weak direct current on Pseudomonas aeruginosa-infected burn wounds. J Trauma. 28(10), 1488–92 (1988).
Kauvar, D. S. et al. Comparison of battlefield-expedient topical antimicrobial agents for the prevention of burn wound sepsis in a rat model. J Burn Care Rehabil. 26(4), 357–61 (2005).
McManus, A. T., McLeod, C. G. Jr. & Mason, A. D. Jr. Experimental Proteus mirabilis burn surface infection. Arch Surg. 117(2), 187–91 (1982).
Mason, A. D. Jr., McManus, A. T. & Pruitt, B. A. Jr. Association of burn mortality and bacteremia. A 25-year review. Arch Surg. 121(9), 1027–31 (1986).
Chan, R. K. et al. Ten years of war: a characterization of craniomaxillofacial injuries incurred during operations Enduring Freedom and Iraqi Freedom. J Trauma Acute Care Surg. 73(6 Suppl 5), S453–8 (2012).
Brandenburg, K. S. et al. Development of Pseudomonas aeruginosa Biofilms in Partial-Thickness Burn Wounds Using a Sprague-Dawley Rat Model. J Burn Care Res. 40(1), 44–57 (2018).
Ma, L. et al. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 188(23), 8213–21 (2006).
Colvin, K. M. et al. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ Microbiol. 14(8), 1913–28 (2012).
Ghafoor, A., Hay, I. D. & Rehm, B. H. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 77(15), 5238–46 (2011).
Ryder, C., Byrd, M. & Wozniak, D. J. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 10(6), 644–8 (2007).
Tettmann, B. et al. Enzyme-Mediated Quenching of the Pseudomonas Quinolone Signal (PQS) Promotes Biofilm Formation of Pseudomonas aeruginosa by Increasing Iron Availability. Front Microbiol. 7, 1978 (2016).
Wiens, J. R. et al. Iron-regulated expression of alginate production, mucoid phenotype, and biofilm formation by Pseudomonas aeruginosa. MBio. 5(1), e01010–13 (2014).
Llamas, M. A. et al. Cell-surface signaling in Pseudomonas: stress responses, iron transport, and pathogenicity. FEMS Microbiol Rev. 38(4), 569–97 (2014).
Taneja, N. et al. Evolution of bacterial flora in burn wounds: key role of environmental disinfection in control of infection. Int J Burns Trauma. 3(2), 102–7 (2013).
Sanjar, F. et al. Temporal shifts in the mycobiome structure and network architecture associated with a rat (Rattus norvegicus) deep partial-thickness cutaneous burn. Med Mycol, https://doi.org/10.1093/mmy/myz030 (2019).
Teplitz, C. et al. Pseudomonas Burn Wound Sepsis. I Pathogenesis of Experimental Pseudomonas Burn Wound Sepsis. J Surg Res. 4, 200–16 (1964).
Friedman, L. & Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 186(14), 4457–65 (2004).
Jackson, K. D. et al. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 186(14), 4466–75 (2004).
Colvin, K. M. et al. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7(1), e1001264 (2011).
Cooley, B. J. et al. The extracellular polysaccharide Pel makes the attachment of P. aeruginosa to surfaces symmetric and short-ranged. Soft Matter. 9(14), 3871–3876 (2013).
Karna, S. L. et al. Genome Sequence of a Virulent Pseudomonas aeruginosa Strain, 12-4-4(59), Isolated from the Blood Culture of a Burn Patient. Genome Announc. 4(2), https://doi.org/10.1128/genomeA.00079-16 (2016).
Kamath, S., Kapatral, V. & Chakrabarty, A. M. Cellular function of elastase in Pseudomonas aeruginosa: role in the cleavage of nucleoside diphosphate kinase and in alginate synthesis. Mol Microbiol. 30(5), 933–41 (1998).
Wretlind, B. & Pavlovskis, O. R. Pseudomonas aeruginosa elastase and its role in pseudomonas infections. Rev Infect Dis. 5(Suppl 5), S998–1004 (1983).
Toder, D. S. et al. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect Immun. 62(4), 1320–7 (1994).
Wolz, C. et al. Pseudomonas aeruginosa LasB mutant constructed by insertional mutagenesis reveals elastolytic activity due to alkaline proteinase and the LasA fragment. Mol Microbiol. 5(9), 2125–31 (1991).
Vasil, M. L. & Ochsner, U. A. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol. 34(3), 399–413 (1999).
Kruczek, C. et al. Serum albumin alters the expression of iron-controlled genes in Pseudomonas aeruginosa. Microbiology. 158(Pt 2), 353–67 (2012).
Baskaran, H., Yarmush, M. L. & Berthiaume, F. Dynamics of tissue neutrophil sequestration after cutaneous burns in rats. J Surg Res. 93(1), 88–96 (2000).
Dong, Y. L. et al. Effect of thermal injury and sepsis on neutrophil function. J Trauma. 34(3), 417–21 (1993).
Tchervenkov, J. I. et al. Early burn wound excision and skin grafting postburn trauma restores in vivo neutrophil delivery to inflammatory lesions. Arch Surg. 123(12), 1477–81 (1988).
Gilpin, D. A. Calculation of a new Meeh constant and experimental determination of burn size. Burns. 22(8), 607–11 (1996).
Leary, S. et al. AVMA Guidelines for the Euthanasia of Animals: 2013 Edition, A.V.M. Association, Editor. American Veterinary Medical Association: Schaumburg, IL. p. 1–102 (2013).
Pirnay, J. P. et al. Quantitation of Pseudomonas aeruginosa in wound biopsy samples: from bacterial culture to rapid ‘real-time’ polymerase chain reaction. Crit Care. 4(4), 255–61 (2000).
Nadkarni, M. A. et al. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 148(Pt 1), 257–66 (2002).
Weaver, A. J. et al. Clinical Utility of PNA-FISH for Burn Wound Diagnostics: A Noninvasive, Culture-Independent Technique for Rapid Identification of Pathogenic Organisms in Burn Wounds. J Burn Care Res. 40(4), 464–470 (2019).
Acknowledgements
The authors would like to acknowledge the help and support of all members of the USAISR DCTR including Ms. Andrea B. Fourcaudot, Ms. Eliza A. Sebastian, and Mr. Andrew Tobias for their help and support in the production of this manuscript. The authors would also like to acknowledge the advice provided by Dr. Ravi Shankar, PhD, regarding development of the burn infection model. Additionally, the authors acknowledge the support of the USAISR Research Support Division for their assistance with animal handling and laboratory support in obtaining systemic neutrophil counts. The study was funded, in part, by the Naval Medical Research Center’s Advanced Medical Development program (MIPR N3239815MHX040) and the Combat Casualty Care Research Directorate, US Army Medical Research and Development Command (USAMRDC). This research was also supported, in part, by an appointment to the Postgraduate Research Participation Program at the U.S. Army Institute of Surgical Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and USAMRDC.
Author information
Authors and Affiliations
Contributions
K.S.B. wrote the main manuscript text, unified the tables/figures for publication, managed the experimental design, performed the animal surgeries and tissue harvesting, and analyzed the data; A.J.W. assisted with the animal surgeries and tissue harvesting, performed the neutrophil counts, and prepared figure 7B; S.L.R.K. performed the quantigene analaysis and prepared figure 6; T.Y. performed the scanning electron microscopy and prepared figure 4; P.C. performed the qPCR and prepared figure 3; L.Q. assisted with the experimental design and performance of the animal surgeries and tissue harvesting; S.V.S. assisted with the qPCR, performed the M.P.O. assay and prepared figure 7A; U.P. assisted with the animal surgeries, animal handling, and producing the gross images of the burn wounds shown in figure 1; J.J.A. prepared the bacterial inoculums, assisted with the quantitative bacteriology, and prepared figure 2; K.P.L. conceived of the presented idea, designed the experimental procedure, and analyzed all the resulting data. All authors reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Brandenburg, K.S., Weaver, A.J., Karna, S.L.R. et al. Formation of Pseudomonas aeruginosa Biofilms in Full-thickness Scald Burn Wounds in Rats. Sci Rep 9, 13627 (2019). https://doi.org/10.1038/s41598-019-50003-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-50003-8
This article is cited by
-
Bacterial biofilm growth and perturbation by serine protease from Bacillus sp.
Archives of Microbiology (2024)
-
Histological assessment, anti-quorum sensing, and anti-biofilm activities of Dioon spinulosum extract: in vitro and in vivo approach
Scientific Reports (2022)
-
Development of an implantable three-dimensional model of a functional pathogenic multispecies biofilm to study infected wounds
Scientific Reports (2022)
-
Burns and biofilms: priority pathogens and in vivo models
npj Biofilms and Microbiomes (2021)
-
Biomimetic Cerium Oxide Loaded Gelatin PCL Nanosystems for Wound Dressing on Cutaneous Care Management of Multidrug-Resistant Bacterial Wound Healing
Journal of Cluster Science (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.