Abstract
IMP-26 was a rare IMP variant with more carbapenem-hydrolyzing activities, which was increasingly reported now in China. This study characterized a transferable multidrug resistance plasmid harboring blaIMP-26 from one Enterobacter cloacae bloodstream isolate in Shanghai and investigated the genetic environment of resistance genes. The isolate was subjected to antimicrobial susceptibility testing and multilocus sequence typing using broth microdilution method, Etest and PCR. The plasmid was analyzed through conjugation experiments, S1-nuclease pulsed-field gel electrophoresis and hybridization. Whole genome sequencing and sequence analysis was conducted for further investigation of the plasmid. E. cloacae RJ702, belonging to ST528 and carrying blaIMP-26, blaDHA-1, qnrB4 and fosA5, was resistant to almost all β-lactams, but susceptible to quinolones and tigecycline. The transconjugant inherited the multidrug resistance. The resistance genes were located on a 329,420-bp IncHI2 conjugative plasmid pIMP26 (ST1 subtype), which contained trhK/trhV, tra, parA and stbA family operon. The blaIMP-26 was arranged following intI1. The blaDHA-1 and qnrB4 cluster was the downstream of ISCR1, same as that in p505108-MDR. The fosA5 cassette was mediated by IS4. This was the first report on complete nucleotide of a blaIMP-26-carrying plasmid in E. cloacae in China. Plasmid pIMP26 hosted high phylogenetic mosaicism, transferability and plasticity.
Similar content being viewed by others
Introduction
Notoriously, extended and overuse of antibiotics have potentiated globally rapid emergence and spread of carbapenem-resistant Enterobacterales (CRE), posing a serious threat to clinical therapy and infection control1,2,3. The major driving force for the diversification and dissemination of CRE has been confirmed as the horizontal transfer of plasmid-mediated carbapenem-hydrolyzing enzymes (i.e., carbapenemase) genes4, among which the most prevalent and of particular clinical importance were blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-485.
IMP, one kind of metallo-β-lactamases (MBLs), can efficiently inactivate almost β-lactams except monobactam5. IMP-1 was the first transferable MBL detected from Pseudomonas aeruginosa in Japan in 19916; subsequently, the continuously clinical detection of blaIMP-1 in different species isolates in Japan7,8, as well as the discovery of IMP-2 in Italy9 and IMP-5 in Portugal10, marked the beginning of the upcoming flourish of IMP MBLs11. IMP-26 was first reported as an IMP-4 variant in Singapore in 2010 from a clinical carbapenem-resistant P. aeruginosa isolate by Koh TH et al.12. However, since then, there have been only sporadic reports on the IMP-26-production in Gram-negative bacilli13,14,15, especially in Enterobacterales15. Notably, isolates expressing IMP-26 were found significantly more resistant to doripenem and meropenem than that expressing IMP-113.
Enterobacter cloacae was one member of the normal intestinal microflora of humans and animals, which has also assumed clinical importance and emerged as a major human pathogen causing hospital-acquired bacteremia, nosocomial pneumonia, urinary tract infections and so on16,17. In the past decade, the emergence of IMP-producing E. cloacae has been extensively reported as a challenge to clinical therapy because of its rapid worldwide transmission14,16,18. And in China, the most common IMP variants found in E. cloacae were IMP-8 and IMP-411,19,20. As for IMP-26-producing E. cloacae, it has been only reported in Chongqing, Shanghai and Beijing worldwide19,21,22.
Our pilot study firstly reported two IMP-26-producing E. cloacae bloodstream isolates in Shanghai21. Considering the higher carbapenem-hydrolyzing activities and emerging reports in China of IMP-26, we subsequently analyzed the transferability and full nucleotide sequence of the corresponding multi-drug-resistance plasmid pIMP26 in this study, which carried several important resistance determinants, such as blaIMP-26, blaDHA-1, aacA4, qnrB4 and fosA5, conferring resistance to carbapenems, cephalosporins, aminoglycoside and fosfomycin, respectively.
Methods
Isolate and antimicrobial susceptibility testing
Isolate RJ702 was obtained from the blood of a female patient with uterine malignancy at Ruijin Hospital in April 2013. The carbapenem-resistant isolate was first isolated at day 28 after admission. The previous travel history of the patient was not documented.
The initial species identification of RJ702 was performed using MALDI-TOF MS (bioMérieux, Marcy-l’Étoile, France). The minimum inhibitory concentrations (MICs) of ceftriaxone, ceftazidime, cefotaxime, cefepime, aztreonam, ciprofloxacin, levofloxacin, amikacin, gentamicin, piperacillin/tazobactam, cefoperazone/sulbactam, trimethoprim/sulfamethoxazole and tigecycline were determined using the broth microdilution method according to guidelines of the Clinical and Laboratory Standards Institute (CLSI M07-A9)23, while that of meropenem, ertapenem and imipenem were determined using the Etest (bioMérieux, Marcy-l’Étoile, France). The susceptibility results were interpreted according to the guidelines of CLSI M100-S2524; while the breakpoint for tigecycline was according to that of European Committee on Antimicrobial Susceptibility Testing (EUCAST) V6.025. Escherichia coli ATCC25922 was used as the quality control. PCR was performed to detect the “big five” carbapenemase genes (blaKPC, blaNDM, blaIMP, blaVIM and blaOXA-48).
Multilocus sequence typing (MLST)
A MLST scheme was used to assign E. cloacae to clonal lineages, including seven housekeeping genes (dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB) as described by Miyoshi-Akiyama26. The combination of seven alleles can define the sequence types (STs) on the MLST website (http://pubmlst.org/ecloacae/).
Plasmid conjugation, S1-nuclease pulsed-field gel electrophoresis (S1-PFGE), and southern hybridization
The transferability of the resistance genes was assessed in broth culture using E. coli J53 Azr (sodiumazide-resistant) as the recipient. The transconjugants were selected on MacConkey agar containing sodiumazide (100 mg/L) and meropenem (2 mg/L) or ceftazidime (1 mg/L). PCR was employed to confirm the existence of blaIMP-26. DNA plugs of the parental and transconjugant digested with S1-nuclease were prepared and separated by PFGE, and then transferred to positively charged nylon membrane (Roche Applied Science, Germany). The membrane was hybridized with digoxigenin-labeled blaIMP-26 specific probes.
DNA sequencing and genomics analysis
Genomic DNA of E. cloacae RJ702 was isolated using ChargeSwitch® gDNA Mini Bacteria Kit (Life Technologies, Carlsbad, CA, USA) and sequenced by a combination of PacBio RSII (Pacific Biosciences, Menlo Park, CA, USA) and Illumina Hiseq X10 (Illumina, San Diego, CA, USA) sequencing platforms. The assembly was produced firstly using a hybrid de novo assembly solution modified by Koren, in which a de-Bruijn based assembly algorithm and a CLR reads correction algorithm were integrated in “PacBioToCA with Celera Assembler” pipeline27,28. The final assembly generated a circular genome sequence with no gap existed. The precise species identification was established based on average nucleotide identity (ANI) between RJ702 and other type strains of E. cloacae subsp. using Orthologous ANI Tool (OAT) recommended by Lee I et al.29. Annotation of the genomic sequence and alignment with other similar sequences were carried out using the BLAST Ring Image Generator (BRIG)30 and SnapGene program v4.3.2. Open reading frames (ORFs) were identified using Glimmer version 3.02 (http://cbcb.umd.edu/software/glimmer/). ORFs less than 300-bp were discarded. Insertion elements and resistance genes were identified using ISFinder (https://www-is.biotoul.fr) and ResFinder (https://cge.cbs.dtu.dk/services/ResFinder). PlasmidFinder (https://cge.cbs.dtu.dk/services/PlasmidFinder) and pMLST (https://cge.cbs.dtu.dk/services/pMLST/) were employed to detect and type the plasmids. BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to identify related plasmids carrying blaIMP to guide PCR-based gap closure and Sanger sequencing to assemble contigs into complete plasmids.
Nucleotide sequence accession number
The completely annotated sequence of pIMP26 in E. cloacae RJ702 has been deposited in GenBank database (Accession Number: MH399264).
Ethics approval and informed consent
The study was approved by Ruijin Hospital Ethics Committee (Shanghai Jiao Tong University School of Medicine), and the Review Board exempted the requirements for informed consent as this study only focused on bacteria.
Results
Precise species identification
RJ702 was initially identified as E. cloacae or E. asburiae by MALDI-TOF MS. Additionally, RJ702 showed 95.2874% ANI value with E. cloacae ECNIH2 (NZ_CP008823) and only 87.8944% with E. asburiae ATCC35953 (NZ_CP011863). The ANI values of RJ702 with E. cloacae subsp. cloacae ATCC13047 (NC_014121) and E. cloacae subsp. dissolvens SDM (NC_018079) were 89.0906% and 87.9528% respectively. Therefore, RJ702 was precisely identified as E. cloacae subsp. cloacae.
Antimicrobial resistance and MLST
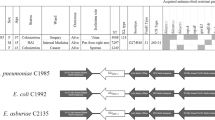
E. cloacae subsp. cloacae RJ702 belonging to ST528, exhibited resistance to cephalosporins, monobactam, carbapenems, β-lactam/β-lactamase inhibitor combinations (only cefoperazone/sulbactam), aminoglycosides, trimethoprim/sulfamethoxazole, and tigecycline. The transconjugant inherited resistance to these antibiotics (Table 1).
Conjugation, S1-PFGE, and southern hybridization
The transconjugant RJ702-1 was obtained by plasmid conjugation experiments. S1-PFGE revealed that RJ702 harbored two plasmids (~320-kb and ~70-kb), and RJ702-1 inherited both. Southern hybridization analysis revealed blaIMP-26 located on the ~320-kb plasmid (pIMP26) (Fig. 1).
Genome sequencing of RJ702
Whole genome sequencing generated 1,458,457 single reads and 4.70 Gb clean data total bases, which were de novo assembled to 184 contigs (75 > 1,000 bp; N50: 269,528 bp; N90: 45,440 bp). The bases in all contigs of RJ702 was 5.01 Mb with a 54.69% G + C content. The size of chromosome was 4,303,224 bp, and the bases in all contigs of two plasmids in RJ702 were 329,420 bp (pIMP26) and 78,322 bp respectively. PlasmidFinder presented that plasmid pIMP26 hosted two replicons, of which IncHI2 was 327 bp and IncHI2A was 630 bp; while the other plasmid hosted none.
Backbones of pIMP26
Plasmid pIMP26 was a 329,420-bp closed circular DNA sequence with an average G + C content of 48.24% (Fig. 2). It hosted two repB replicons, and both belonged to IncHI2 prototype (ST1 subtype). BLAST searches indicated the backbone regions of pIMP26 highly similar to a 324,503-bp IncHI2 plasmid pEC-IMPQ (Genbank ID: EU855788) from an IMP-8-producing E. cloacae isolate in Taiwan (87% query coverage and 99% nucleotide identity) (Fig. 2). Annotation of the finished sequence data revealed that pIMP26 contained 381 ORFs, including repB (position: 32558–33613 and 47056–47931, for plasmid replication initiation), trhK/trhV (for stabilizing mating pairs during plasmid conjugation), four transfer gene clusters (locus traB, traN, traG and traM, encoding the conjugative apparatus), the parA family operon (for the replication/partitioning system with repB), and the stbA family operon (for plasmid stability).
Circular map of plasmid pIMP26. The two inner circles represented the G + C content plotted against the average G + C content of 48.24% (black circle) and GC skew information (green and purple circles). Circles in different colors represented different plasmids (details in the legend), and the Genbank numbers were as follows: pIMP26 (MH399264), p505108-MDR (KY978628), pCNR48 (LT994835), pEC-IMPQ (EU855788), pIMP4-SEM1 (FJ384365), and pGMI14-002 (CP028197). The location of discussed resistance genes and intI were also demonstrated on the outer cyan-blue circle. The annotation of the genetic components were added manually using the Microsoft PowerPoint 2016 program.
Resistance regions in pIMP26
Plasmid pIMP26 was rich in mobile genetic elements, including IS elements (IS4, IS6, IS26, ISCR1, etc.), transposons (Tn3 family, etc.) and integrons (intI1, etc.), and contained multiple resistance genes (blaIMP-26, fosA5, blaDHA-1, qnrB4, aac(6′)-Ib3, aac(6′)-IIc, aacA4, aph(6)-Id, strA, mph(A), ere(A), catA2, tet(D), dfrA18, blaSHV-12, two copies of blaTEM-1B, and three copies of sul1). According to BLAST searches (Fig. 3), the blaIMP-26 region was sequentially arranged as intI1, blaIMP-26, ltrA, qacEΔ1 and sul1 (position: 2567–7765), same as the blaIMP-4 cluster in pIMP-4 from an IMP-4-producing Klebsiella pneumoniae isolate in Shanghai (Genbank ID: FJ384365). In pIMP26, blaIMP-26 cassette is the downstream of IS6 and followed by IS6100 (IS6-like), Eco128I (type II restriction enzyme) and M. EcoRII (type II methyltransferase). The fosA5 cluster was arranged sequentially as IS4, RfaY, LysR, fosA5, RfaY, ISVsa5 (IS4-like) (position: 13522–20464). The upper half of the cluster was opposite to that of pHKU1 from a fosA5-producing E. coli isolate in HK (Genbank ID: KC960485). The blaDHA-1 and qnrB4 cluster was sequentially arranged as ISCR1, sapC, sapB, sapA, YdeJ, qnrB4, pspE, pspA, pspB, pspC, pspD, LacI, blaDHA-1, LysR, hypA, qacEΔ1 and sul1 (position: 169330–194847). It shared high similarity with p505108-MDR from a blaDHA-1- and qnrB4-producing Cronobacter sakazakii in China (Genbank ID: KY978628). Besides, two copies of blaTEM-1B were both located in Tn3 transposon in pIMP26.
Plasmid accessory resistance regions. The comparison of linear DNA against the corresponding regions in different plasmids. The resistance genes were indicated by orange arrows and the insertion sequences are indicated by purple arrows. Shading regions denoted regions of homologous (>95% nucleotide identity).
Discussion
The undesirable antibiotic resistance (especially carbapenem-resistance) has appeared and disseminated rapidly in Gram-negative bacilli, which was attributed largely to the acquisition of multiple resistance genes by horizontal plasmid-mediated genes transfer4,5. Our study was to map the genetic environment of a novel multi-drug-resistance plasmid pIMP26, in order to provide a new insight for the potential spread of blaIMP-26 and fosA5 or correlations between genetic diagnosis and clinical treatment.
Firstly, the backbone of pIMP26 was blasted with different plasmids in BLAST. The origins of functional modules in pIMP26, such as multiple antibiotic resistance determinants, stably conjugal transfer (tra and trh family), mobile elements and plasmid maintenance (stb family) (Fig. 2), represented a strong transferability, stability and plasticity of this plasmid31. IncHI2 was one of the most prevalent broad-host-range plasmid families carrying different resistance determinants simultaneously in Enterobacterales4,31,32. As previously reported on E. cloacae, most β-lactamase-encoding genes (blaSHV-12, blaCTX-M-15, blaNDM-1, blaIMP-4, etc.) were also located on IncHI2 plasmids (subtype ST1) of 290~340-kb in size18,31,32,33,34,35, and our study also fit it. It should be noted that the similar backbone shared by pIMP26 and other plasmids (Fig. 2) in clinical isolates of E. cloacae, K. pneumoniae and S. enterica from different areas strongly suggested that inter-species genetic exchange also occurred, thus broadening the host range and dissemination of combined cargo genes. Besides, pIMP26 contained a wide variety of transposable elements carrying known antibiotic resistance genes. Tn3 family transposon was the medium of TEM genes and fosA5 was also located in Tn3 in pIMP26 (Figs 2 and 3). The archetype of Tn3 was known as some of the earliest unit transposons identified in Gram-negative bacilli. Tn3 family members demonstrated transposition immunity, but homologous and/or res-mediated recombination between related elements can occur, creating hybrid elements31. And this would explain multiple Tn3-mediated resistance elements in pIMP26 in this study. However, further study is definitely needed to characterize the mechanisms behind the transfer or recombination of Tn3.
IMP-26, firstly found in P. aeruginosa in Singapore, was differed from IMP-4 at position 145 (G to T change); the translated amino acid sequence differed from IMP-4 at residue 49 (phenylalanine for valine)12. Blast searches indicated that the genetic structure surrounding blaIMP-26 has only revealed in a study from Vietnam up to now, containing intI1-blaIMP-26-qacG-aac(6′)-Ib-orf3-orf4 (Fig. 2)13, and our study was the first time focusing on the complete nucleotide of the plasmid carrying blaIMP-26. Interesting was the blaIMP-26 region in pIMP26 different from that found in Vietnam (though both located on intI1)13; but same as the blaIMP-4 cluster of pIMP-4 in Shanghai (Genbank ID: FJ384365) (Fig. 3). It prompted that the blaIMP-26 detected in our study maybe originated from blaIMP-4 or the genetic mutation may occur during transfer of blaIMP-26 cassette.
The prevalence and dissemination of fosA5 have probably been underestimated36. Previous study once found IS10 playing an important role in the mobilization of fosA537. However, the upper half consistent with pHKU1 in pIMP26 indicated that IS4 might also related to its mobilization36 (Fig. 3). Plasmid carrying blaDHA was usually reported also carrying qnrB4, blaSHV-1238. This suggested that the cassette in common of qnrB4 and blaDHA-1 (Fig. 3) (including that in pIMP26) was derived from the same immediate ancestor. The qnrB4-blaDHA-containing region of pIMP26 was located after the 3′ conserved sequence (3′-CS) of intI1 (Fig. 3), containing aac(6′)-IIc, qacEΔ1 and sul1. Besides, an insertion sequence common region 1 (ISCR1) was identified downstream of sul1. ISCR1 could mobilize the nearby sequence and a truncated 3′-CS from one integron to the 3′-CS of another integron through rolling-circle transposition, and provide a promoter for the expression of nearby genes39; this may lead to the co-carriage of multiple resistant genes in one plasmid and the multi-drug resistance of clinical isolates.
Interestingly, our study showed the qnrB4- and aac(6′)-Ib3-harboring RJ702 susceptible to quinolones (MIC = 0.5 or 0.25). We speculated that it was due to the absence of other mechanisms of chromosomal resistance (e.g. alterations in type II topoisomerases) in RJ702 other than plasmid-mediated quinolone resistant (PMQR) genes. Researchers found that PMQR mechanism caused only low-level quinolone-resistance on its own, which may not exceed the clinical breakpoints of susceptibility for quinolones but facilitated selections of higher-level resistance and posed threats to the treatment of infections by microorganisms hosting PMQR genes40, which could validate our speculation and underline the necessity of monitoring on PMQR genes.
This is the first report on the entire structure of blaIMP-26-carring plasmid. To some extent, our study evidenced the increasing clinical significance of IncHI2 replicons as resistance genes’ reservoirs and provided insights on the possibilities of further spread in China and highlighted the needs for intensive surveillance and precautions.
Conclusions
We firstly reported here the complete nucleotide sequence of a plasmid carrying blaIMP-26, which was an IncHI2 replicon simultaneously encoding multidrug resistance determinants, including β-lactam (blaIMP-26, blaDHA-1, blaSHV-12, etc.), aminoglycoside (aac(6′)-IIc, aacA4, aph(6)-Id, etc.), fluoroquinolone (qnrB4, aac(6′)-Ib3) and fosfomycin (fosA5) resistance genes. New genetic context of fosA5 was also characterized. The novel plasmid with multi-insertion of different resistant components and stable inheritance emphasized controlled use of clinical antibiotics to prevent selective pressure aggravating the emergence and dissemination of multi-drug resistance.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Kaier, K., Frank, U., Hagist, C., Conrad, A. & Meyer, E. The impact of antimicrobial drug consumption and alcohol-based hand rub use on the emergence and spread of extended-spectrum beta-lactamase-producing strains: a time-series analysis. The Journal of antimicrobial chemotherapy 63, 609–614 (2009).
Jean, S. S. & Hsueh, P. R. High burden of antimicrobial resistance in Asia. International journal of antimicrobial agents 37, 291–295 (2011).
Wang, S. et al. Changes in antimicrobial susceptibility of commonly clinically significant isolates before and after the interventions on surgical prophylactic antibiotics (SPAs) in Shanghai. Brazilian Journal of Microbiology 49, 552–558 (2018).
Carattoli, A. Plasmids and the spread of resistance. International journal of medical microbiology: IJMM 303, 298–304 (2013).
Bush, K. Carbapenemases: Partners in crime. Journal of global antimicrobial resistance 1, 7–16 (2013).
Watanabe, M., Iyobe, S., Inoue, M. & Mitsuhashi, S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrobial agents and chemotherapy 35, 147–151 (1991).
Osano, E. et al. Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrobial agents and chemotherapy 38, 71–78 (1994).
Senda, K. et al. PCR detection of metallo-beta-lactamase gene (bla IMP) in gram-negative rods resistant to broad-spectrum beta-lactams. Journal of clinical microbiology 34, 2909–2913 (1996).
Cornaglia, G. et al. Appearance of IMP-1 metallo-beta-lactamase in Europe. Lancet (London, England) 353, 899–900 (1999).
Da Silva, G. J. et al. Molecular characterization of bla(IMP-5), a new integron-borne metallo-beta-lactamase gene from an Acinetobacter baumannii nosocomial isolate in Portugal. FEMS microbiology letters 215, 33–39 (2002).
Mojica, M. F., Bonomo, R. A. & Fast, W. B1-Metallo-beta-Lactamases: Where Do We Stand? Current drug targets 17, 1029–1050 (2016).
Koh, T. H. et al. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-beta-lactamase genes, including the novel bla(IMP-26) gene. Journal of clinical microbiology 48, 2563–2564 (2010).
Tada, T. et al. Multidrug-Resistant Sequence Type 235 Pseudomonas aeruginosa Clinical Isolates Producing IMP-26 with Increased Carbapenem-Hydrolyzing Activities in Vietnam. Antimicrobial agents and chemotherapy 60, 6853–6858 (2016).
Peirano, G., Lascols, C., Hackel, M., Hoban, D. J. & Pitout, J. D. Molecular epidemiology of Enterobacteriaceae that produce VIMs and IMPs from the SMART surveillance program. Diagnostic microbiology and infectious disease 78, 277–281 (2014).
Huang, S., Dai, W., Sun, S., Zhang, X. & Zhang, L. Prevalence of plasmid-mediated quinolone resistance and aminoglycoside resistance determinants among carbapeneme non-susceptible Enterobacter cloacae. PloS one 7, e47636 (2012).
Mezzatesta, M. L., Gona, F. & Stefani, S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol 7, 887–902 (2012).
Davin-Regli, A. & Pages, J. M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Frontiers in microbiology 6, 392 (2015).
Sidjabat, H. E., Heney, C., George, N. M., Nimmo, G. R. & Paterson, D. L. Interspecies transfer of bla IMP-4 in a patient with prolonged colonization by IMP-4-producing Enterobacteriaceae. Journal of clinical microbiology 52, 3816–3818 (2014).
Dai, W. et al. Characterization of carbapenemases, extended spectrum beta-lactamases and molecular epidemiology of carbapenem-non-susceptible Enterobacter cloacae in a Chinese hospital in Chongqing. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases 14, 1–7 (2013).
Feng, W. et al. Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Scientific Reports 6, 33419 (2016).
Wang, S. et al. Antimicrobial susceptibility and molecular epidemiology of clinical Enterobacter cloacae bloodstream isolates in Shanghai, China. PloS one 12, e0189713 (2017).
Jin, C. et al. Molecular Characterization of Carbapenem-Resistant Enterobacter cloacae in 11 Chinese Cities. Frontiers in microbiology 9, 1597 (2018).
Clinical and Laboratory Standards Institute (CLSI). Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; Approved standard-9th edition. CLSI document M07–A9 (2015).
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI document M100–S25 (2015).
European Committee On Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 6.0., http://www.eucast.org/clinical_breakpoints/ (2016).
Miyoshi-Akiyama, T., Hayakawa, K., Ohmagari, N., Shimojima, M. & Kirikae, T. Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PloS one 8, e66358 (2013).
Chin, C. S. et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature methods 10, 563–569 (2013).
Koren, S. et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nature biotechnology 30, 693–700 (2012).
Lee, I., Ouk Kim, Y., Park, S. C. & Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. International journal of systematic and evolutionary microbiology 66, 1100–1103 (2016).
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L. & Beatson, S. A. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (2011).
Partridge, S. R., Kwong, S. M., Firth, N. & Jensen, S. O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clinical microbiology reviews 31, e00088–00017 (2018).
Coelho, A. et al. Role of IncHI2 plasmids harbouring bla VIM-1, bla CTX-M-9, aac(6′)-Ib and qnrA genes in the spread of multiresistant Enterobacter cloacae and Klebsiella pneumoniae strains in different units at Hospital Vall d’Hebron, Barcelona, Spain. International journal of antimicrobial agents 39, 514–517 (2012).
Nilsen, E. et al. Large IncHI2-plasmids encode extended-spectrum beta-lactamases (ESBLs) in Enterobacter spp. bloodstream isolates, and support ESBL-transfer to Escherichia coli. Clinical microbiology and infection: the official publication of the European Society of Clinical Microbiology and Infectious Diseases 19, E516–518 (2013).
Haenni, M. et al. High prevalence of international ESBL CTX-M-15-producing Enterobacter cloacae ST114 clone in animals. The Journal of antimicrobial chemotherapy 71, 1497–1500 (2016).
Petrosillo, N. et al. Spread of Enterobacter cloacae carrying bla NDM-1, bla CTX-M-15, bla SHV-12 and plasmid-mediated quinolone resistance genes in a surgical intensive care unit in Croatia. Journal of global antimicrobial resistance 4, 44–48 (2016).
Ho, P. L. et al. Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. Journal of medical microbiology 62, 1707–1713 (2013).
Ma, Y. et al. Characterization of fosA5, a new plasmid-mediated fosfomycin resistance gene in Escherichia coli. Letters in applied microbiology 60, 259–264 (2015).
Hennequin, C., Ravet, V. & Robin, F. Plasmids carrying DHA-1 beta-lactamases. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology 37, 1197–1209 (2018).
Chen, Y. T. et al. Mobilization of qnrB2 and ISCR1 in Plasmids. Antimicrobial agents and chemotherapy 53, 1235–1237 (2008).
Rodriguez-Martinez, J. M. et al. Plasmid-mediated quinolone resistance: Two decades on. Drug Resist Updat 29, 13–29 (2016).
Acknowledgements
We would like to thank the Department of Medical Microbiology and Parasitology at the School of Medicine of Shanghai Jiao Tong University for excellent laboratory provision and technical assistance. This work was supported by the National Natural Science Foundation of China (Grant No. 81772245), the Shanghai Three-Year Plan of the Key Subjects Construction in Public Health-Infectious Diseases and Pathogenic Microorganism (Grant No. 15GWZK0102) and the Special Fund for Health-scientific Research in the Public Interest of China Program (grant no. 201002021). The funders had no role in study design, data collection and analysis, preparation of the manuscript or decision for publication.
Author information
Authors and Affiliations
Contributions
Conception and design of study: L.H. and J.S., S.W. Acquisition of data (laboratory or clinical): S.W., K.Z., S.X., F.G. and X.L. Analysis and interpretation of data: S.W., K.Z. and L.X. Contribution of reagents/materials/analysis tools: L.H. and Y.N. Drafting of article and/or critical revision: S.W., K.Z., L.H. and J.S. Final approval of the submitted manuscript: All.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Zhou, K., Xiao, S. et al. A Multidrug Resistance Plasmid pIMP26, Carrying blaIMP-26, fosA5, blaDHA-1, and qnrB4 in Enterobacter cloacae. Sci Rep 9, 10212 (2019). https://doi.org/10.1038/s41598-019-46777-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-46777-6
This article is cited by
-
Outbreak of multi-drug-resistant (MDR) Shigella flexneri in northern Australia due to an endemic regional clone acquiring an IncFII plasmid
European Journal of Clinical Microbiology & Infectious Diseases (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.