Abstract
Regional gender differences in autosomal chromosome disorders have been observed repeatedly. However, the corresponding diversity changes remain unconfirmed. By analyzing previously published thalassemia data from the Dai people in Dehong and Xishuangbanna (two regions in Yunnan Province, China), we found that several sequence types, including HBA CNV and HBB mutations, significantly depend on gender in Xishuangbanna but not in Dehong. With the supportive evidence from previous researches, we accept that some certain mutations depend on gender regionally. This association seems peculiar. It is among one common people on a small geographical scale, while other recorded thalassemia gender difference varies by ethnics and continent.
Similar content being viewed by others
Introduction
In past decades, various gender differences in autosomal chromosome disorders have been observed1,2,3,4,5,6, and corresponding differences in related mutation frequencies were found7,8. However, the regionality of these genetic gender dependencies has never been investigated, even though some gender differences in autosomal disorders are regional9,10,11,12,13,14,15. Thus, we aimed to determine the regional gender dependency of genetic factors.
We focused on thalassemia among the Dai people in Dehong and Xishuangbanna (two regions in Yunnan Province, China).
Thalassemia is an autosomal chromosome disorder16 (https://en.wikipedia.org/wiki/Thalassemia) with various gender differences4,5,6,9,10,11,12,13,14,15. The severity of the bone disease and cardiac disorder associated with thalassemia varies by gender4,5,6. A significant association between gender and alloimmunization during transfusion was found in African Americans9 but not in Indians10,11. Similar phenomena were found for selection due to malaria. In Kenya, malaria risk is higher for females than for males, and in India, this pattern is reversed12,13. However, females in India are more likely to die from severe malaria, even if differences in therapy between genders are eliminated; this likelihood is not observed in Ethiopia14,15. The cases above involve gender-dependent fitness, potential mortality and regional selection. These effects could result in a regional diversity difference between genders.
Dehong and Xishuangbanna are located in the southern part of Yunnan Province, China. Both locations have a hot climate year round, with rainy weather during the wet season. The inhabitants are at high risk of malaria17,18 and have a high incidence of thalassemia19,20,21. According to studies performed in the 1980s and 2010s, abnormal hemoglobin was found to be frequent in various minorities21,22. Additionally, a genetic test in 2011 revealed that the carrier rates for pathogenic thalassemia mutations among minority children were 46% in Dehong and 40% in Xishuangbanna23. This result was supported by another genetic test in 201324.
The Dai people are populous in Dehong and Xishuangbanna and have an extraordinarily high frequency of pathogenic thalassemia mutations25,26. Although the two regions are located close to each other and have almost the same ethnic composition, neglected regional gender differences were found when we interpreted Yao’s work27 (see the supplement for our interpretation of previous research). The beta thalassemia incidence among Dai children (by a hemoglobin test) depends significantly on gender in three regions of Xishuangbanna, but not in any observed region of Dehong.
Our work is based on a previous study that was published in 201728. In this study, 951 Dai individuals from Dehong and Xishuangbanna were tested with next-generation DNA sequencing, and basic personal information was recorded (see DataForAna in S1_Table.xls). We mined the data and compared the neglected association factors of sequence type frequency in two regions. If the frequencies are related to gender, we will accept the hypothesis that thalassemia gender differences result in consequent diversity differences between genders. If some of these relationships are significant in only one region, we will conclude that the gender-dependent diversity difference is regional.
Results
The frequency of each sequence type is shown in Fig. 1 (Fig. 1A–E, see CompareTypeFrequency01 in S1_Table.xls). A total of 31 sequence types were recorded. Most sequence types had a frequency of less than 5%, and several sequence types appeared to differ between Xishuangbanna and Dehong. By Fisher’s exact test, the differences in six sequences types were found to be significant; these sequence types include three hemoglobin alpha copy number variations (“αα/−α3.7”, “αα/−−SEA”, and “αα/−α4.2”) and three hemoglobin beta mutations (“HBB: c.79G > A(het)”, “HBB: c.126_129delCTTT(het)”, and “HBB: c.52A > T(het)”) (Fig. 1A–E). Although the sequence type number suggested that the number of combinations of these sequence types would be large (Fig. 1A–E, see CompareTypeFrequency01 in S1_Table.xls), only 53 genotypes were observed. The frequencies of nine genotypes differed significantly between the two regions and are shown in Fig. 1 (Fig. 1F, see CompareTypeFrequency02 in S1_Table.xls).
Diversity comparison. In panels A-E, the frequency of each sequence type is presented. “*” indicates that the difference is significant. Panel F shows that all the genotypes differed significantly between the two regions. The paired code name and type annotation listed in the legend for each genotype was used in the rest of this article, including in the description of genotypes and the identification of genotypes for association rules. (A) HBA1: hemoglobin, alpha 1. A100 = normal, A101 = c.95 + 1G > A, A102 = c.98T > A. (B) HBA2: hemoglobin, alpha 2. A200 = normal, A201 = c.427T > C (het), A202 = c.369C > G (het), A203 = c.369C > G (hom), A204 = c.1delA (hom), A205 = c.427T > C (hom), A206 = c.377T > C (hom). (C) HBA_CNV: hemoglobin, alpha, copy number variation. AC00 = normal, AC01 = αα/−α3.7, AC02 = αα/ −α3.7(α1 and α2 fusion after deletion in another chain), AC03 = αα/–SEA, AC04 = αα/−α4.2, AC05 = −α3.7/= SEA, AC06 = −α3.7/−α3.7, AC07 = αα/ααα. (D) HBB: hemoglobin, beta. B00 = normal, B01 = c.79G > A (het), B02 = c.217dupA (het), B03 = c.126_129delCTTT (het), B04 = c.52A > T (het), B05 = c.316-238C > T (het), B06 = c.79G > A (hom), B07 = c.380T > G (het), B08 = c.410G > A (het), B09 = c.126delC (het), B10 = c.92 + 1G > T (het). (E) HBB_CNV: hemoglobin, beta, copy number variation. BC00 = normal, BC01 = 1-HBB. Here, “het” and “hom” denote heterozygote and homozygote. The details of the code names can be seen in S2_Table.xls. (F) Each genotype is a combination of sequence types, and every combination is described with the involved sequence types. Genotype 1 = A100, A203, AC03, B00, BC00; Genotype 2 = A100, A202, AC00, B00, BC00; Genotype 3 = A100, A200, AC04, B00, BC00; Genotype 4 = A100, A200, AC03, B00, BC00; Genotype 5 = A100, A200, AC01, B01, BC00; Genotype 6 = A100, A200, AC01, B00, BC00; Genotype 7 = A100, A200, AC00, B04, BC00; Genotype 8 = A100, A200, AC00, B03, BC00; Genotype 9 = A100, A200, AC00, B01, BC00.
Discounting redundant rules, 181 and 161 association rules were found in Xishuangbanna and in Dehong, respectively (see AssociationRulesBN, AssociationRulesDH in S1_Table.xls). In both regions, the rules with the highest lift all had support values less than 0.1, and in most other cases, the lift was near 1 (Fig. 2, see AssociationRulesBN, AssociationRulesDH in S1_Table.xls). In each region, the rules are divided into 20 groups by the left-hand side (LHS) of the implication (Fig. 2). The rule classification shows that the rules identified by the apriori algorithm are regional in a sense. In Xishuangbanna, the rules with the highest lift always have an LHS containing “HBA_CNV = AC03, Sex = Female” or “Sex = Male, Age = O” (AC03 is for “αα/ −− SEA”). In Dehong, the lifts are highest when the LHS contains “HBB = B01” and “sex = Female” (B01 is for “HBB: c.79G > A(het))”.
Scatter diagram and classification diagram for association rules. The support, evidence and lift in the scatter diagrams were given by the apriori algorithm and are the most important properties of an association rule. The high lift indicates that the identified rule cannot be caused by randomness. In the classification diagrams, the association rules were classified by the left-hand side (LHS) of the implication. Rules that share more common property-value pairs in the LHS are thought to be more similar.
Most of the associations were not accepted (see Method). According to the Fisher’s exact test (see FTestAssociationRulesBN, FTestAssociationRulesDH in S1_Table.xls), the odds ratios (see Method) for 12 rules from Xishuangbanna departed from 1 significantly, and these rules were thought to be acceptable (Fig. 3, see FTestAssociationRulesBN in S1_Table.xls). In contrast, no rules from Dehong had an odds ratio that significantly departed from 1, although two rules had p values of 0.0504 and 0.07 (see FTestAssociationRulesDH in S1_Table.xls). These p values could be due to the rarity of the relevant sequence types, and these rules might be judged to be significant in future studies with a much larger sample size, but they were not considered significant here.
Probability comparison for significant association rules. Each pair of bars represents a significant rule. This figure shows the difference between the joint probability and the conditional probability corresponding to each significant association rule. The joint probability is described by the percentage of the samples corresponding to the right-hand side (RHS) of the rule, which indicates the frequency in all samples. The conditional probability is described by the percentage of the samples corresponding to both the LHS and the RHS (left- and right-hand sides of the rule), which indicates the frequency according to the rule. Rule 1 {Sex = Female, HBA_CNV = AC03, HBB = B00} => {HBA2 = A203}, Rule 2 {HBA2 = A203} => {HBA_CNV = AC03}, Rule 3 {Sex = Male, Age = Y} => {HBB = B01}, Rule 4 {Sex = Male, HBA2 = A200, HBA_CNV = AC00} => {HBB = B04}, Rule 5 {Sex = Male, Age = O, HBA2 = A200, HBB = B00} => {HBA_CNV = AC03}, Rule 6 {Sex = Male, Age = O, HBB = B00} => {HBA_CNV = AC03}, Rule 7 {Sex = Male, Age = O, HBA2 = A200} => {HBA_CNV = AC03}, Rule 8 {Sex = Male, HBA2 = A200, HBB = B00} => {HBA_CNV = AC03}, Rule 9 {Sex = Male, Age = O} => {HBA_CNV = AC03}, Rule 10 {Sex = Male, HBB = B00} => {HBA_CNV = AC03}, Rule 11 {HBA2 = A200, HBA_CNV = AC03} => {Sex = Male}, Rule 12 {Sex = Male} => {HBA_CNV = AC03}. The significant association rules found in Xishuangbanna were divided into three groups here.
The first group relates to the combination of certain pathogenic mutation types. Only one rule is in this group, “{HBA2 = A203} => {HBA_CNV = AC03}”. This rule holds that a person with the mutation “HBA2: c.369C > G (hom)” (code with “A203”) is more likely to carry “αα/−−SEA” (coded with “AC03”).
The second group suggests an association between being female and having certain sequence type combinations. This group also contains only one rule, “{Sex = Female, HBA_CNV = AC03, HBB = B00} => {HBA2 = A203}”. This rule holds that with the pathogenic mutation “αα/−−SEA” (coded with “AC03”) and normal HBB, a female is more likely to carry “HBA2: c.369C > G (hom)” (coded with “A203”).
The third group indicates an association among male sex, age and certain combinations of sequence types. This group has nine rules, all with “Sex = Male” (Fig. 3, rules 3–11). This group can be summarized with a few knowledge points. First, the frequency of “HBB: c.79G > A(het)” (coded with “B01”) is higher among young men (see “{Sex = Male, Age = Y} => {HBB = B01}”). Second, when both HBA2 and HBA CNV are normal, a higher frequency of “HBB: c.52A > T(het)” (coded with “B04”) can be expected in men (see “{Sex = Male, HBA2 = A200, HBA_CNV = AC00} => {HBB = B04}”). The rest of the rules (Fig. 4, rules 5–12) in this group can be summed as the possibility that “αα/−−SEA” varies among men according to HBA2, HBB and age.
Data analysis process. The process includes multiple steps: data cleaning, association mining, association rule significance testing, association regionality discussion, and association rule replacement discussion. The data were cleaned mainly by removing the items without needed information. The associations were mined with the apriori algorithm. The association rules identified by apriori were tested with the Fisher’s exact test. Then, the regionality and replacement of the significant rules were discussed. When discussing regionality, each significant rule found in a region was tested in the other region. If a rule was significant in one region but not in the other, it was thought to be regional. When discussing the rationality of replacing an apriori rule with a simpler rule, a comparison was made between two conditional probabilities, one for the apriori rule and the other for a given simpler rule that could replace the former rule. If the probabilities differed from each other significantly, the replacement was thought not to be rational. At last, a permutation test was designed for the Xishuangbanna rules including “Sex” in discussion.
The rules that were significant in Xishuangbanna (Fig. 3) were tested in Dehong. The results of these additional Fisher’s exact tests suggested that no rule is significant in both region (see RuleBNInDH in S1_Table.xls), since most p values were near 1 in Dehong. It was accepted that the significant rules found in Xishuangbanna are regional.
A conditional probability comparison was used to discuss whether a given simpler rule can serve as a replacement for a rule produced by the apriori function (see ComparisonBN in S1_Table.xls). The Fisher’s exact test outputted the p values for 21 comparisons that were less than or equal to 0.05 (Table 1.). This outcome means that these rules identified by apriori cannot be replaced with the given simpler rules. However, other replacements (see ComparisonBN in S1_Table.xls) may be rational.
Based on the 21 comparisons (Table 1), gender and “αα/−−SEA” (coded with “AC03”) seem essential. When removing any one of these parameters from the LHS of an apriori rule, the p value for the comparison is less than 0.05.
Discussion
Although 12 BN rules were listed in Fig. 3, only three rules, {Sex = Male} => {HBA_CNV = AC03}, {Sex = Male, Age = O} => {HBA_CNV = AC03}, and {Sex = Male, Age = Y} => {HBB = B01}, were seriously discussed here, that because they are more credible. They do not include rare mutations which are not able to be well researched with a small data set. Additionally, these rules had been confirmed by permutation test with shuffling “Sex”.
According to these three rules, the frequencies of certain thalassemia mutations depend on gender in Xishuangbanna but not in Dehong. This dependence had never been reported in Dai people from Xishuangbanna, because no genetic data set had been got from them before 2016. However, the three rules are consistent with previous researches27,29.
The rules {Sex = Male} => {HBA_CNV = AC03}, {Sex = Male, Age = O} => {HBA_CNV = AC03} are supported by malaria incidence record29. These two rules mean that male, especially elder male have more chance to carry mutation “αα/−−SEA”, which was selected by malaria in China30. The malaria record is from 1981 to 201029. It can be briefly described as the following points:
-
a.
In Xishuangbanna, the cumulative malaria incidence from 1981 to 2005 was about 7.5%, that is considerable.
-
b.
In Xishuangbanna, only very few malaria incidences were found after 2006.
-
c.
In Xishuangbanna, men in the age between 15 and 35 are with the highest malaria risk. Thus, the men’s cumulative incidence from 1981 to 2005 should be greater than 7.5%, which is based on all people, including men and women.
In our research, elder men were (defined as) born since mid 1970s to 1989 (elder than 24 in 2013); younger men were born after 1990. According to their birth year, it is reasonable to accept that, the elder men had been affected much more than younger.
The elder group were older than 15 years at year 2006. When the malaria incidence was considerable (a), they were at the age of highest risk (c). In this group, the men’s cumulative incidence from 1981 to 2005 should be considerable (c). Even so, the incidence might be underestimated. Because the elder men born in mid 1970s did not count the incidence rate, the incidences were not recorded by medical workers at that time.
Comparatively, the younger had much less infection opportunities: before 2005, they were under 15 years old, the record showed that the children of this age were at much lower risk (c); and after 2006, medical prevention made incidences very rare (b) when they were old enough to get high risk.
By the interpretation above, we found a perfect correspondence: more frequent antimalaria mutation “αα/––SEA” are among male, especially elder male (according to our rules), exactly the people at highest risk of infection (according to the previous record). This correspondence suggests that, malaria caused selection can make the observed association.
The rule, {Sex = Male, Age = Y} => {HBB = B01}, is consistent with the a detailed anemia investigation27. In this investigation, it was showed that beta thalassemia depends on gender among Dai children in Xishuangbanna, while not in Dehong27. Considering ‘B01’ is one of the most frequent beta thalassemia mutation type, we conclude that {Sex = Male, Age = Y} => {HBB = B01} might be a cause of the previously observed gender dependence. Moreover, our rule and previously finding on gender dependence27 have common regionality: it is not found in Dehong. The cause of this regionality still remains unknown. The exploration of the cause requires more researches on the mechanism.
Considering the consistence with previous researches, we accept that, certain thalassemia mutations depend on gender regionally. Additionally, it is noteworthy that our identified association rule is also a regional gender difference among one native people on a small geographical scale (Dehong and Xishuangbanna are approximately 400 km from each other). In contrast, most thalassemia gender differences vary by continent or ethnicity7,8,9,10,11. We believe that this abnormal regionality results from other neglected factors, such as social life, isolation or unknown selection mechanism. Moreover, the regional gender dependence indicates an interesting question: the likelihood of passing an autosomal genetic disorder to offspring might rely on gender regionally. This likelihood proposed by us is hypothetical and also remains unexplored.
Because our study is based on the first genetic data set for Dai people in Xishuangbanna and Dehong, it is the first data mining, an exploratory work. Considering only 35 males and more than 200 females in the Xishuangbanna set, we tried our best to avoid biased conclusion: only the frequent mutations were considered; the conclusions were drawn on the consideration of the supportive evidences from previous researches, of which sample size ranged from hundreds to thousands27,29, much bigger than ours. Actually, our conclusion is not based on a single small sample size. The impact by the imperfections of the data set was eliminated as possible. Of course, better data set is necessary for further work.
Methods
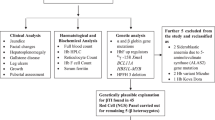
Before analysis, less than twenty samples were removed because of the absence of basic personal information or genetic test results that were needed for subsequent data mining. Ultimately, 264 samples from Xishuangbanna and 664 samples from Dehong were in the analyzed set (see DataForAna in S1_Table.xls).
The first analysis is a comparison study. The frequency of each sequence type (including copy number variation) was compared between two regions. The frequencies of the observed combinations of these types were also compared. Significance was tested with Fisher’s exact test.
The analysis process for association factors is shown in Fig. 4.
The most important analysis is the association between gender and mutation. Only two genes were recorded in the studied data set. In similar contexts, each mutation is usually studied individually31. However, when too many mutation types are involved, it is difficult to list all possible associations manually. Thus, association mining was used to generate the hypothesis here. The importance of each outputted association was evaluated by significance tests and previous studies, as in research methods that focus on a few genes31, because Bonferroni adjustment cannot apply to such associations. The occurrence of these mutation types is not independent. In most cases, the occurrence of a mutation means that other mutation types are not found in the same gene.
The function ‘apriori’ in the R package ‘aRules’ was used to search for possible associations. Before running the function apriori, the age of each person was converted to a factor that can be considered by the apriori algorithm. Ages greater than the median age were considered old (labeled ‘O’), and ages less than or equal to the median were considered young (labeled ‘Y’). Thereafter, if an association rule involved age, the corresponding logical implication was indicated with an ‘O’ or ‘Y’. Counting the pathogenic mutation types with very low frequency, the parameters of the function apriori were set manually. The parameter ‘support’ was set to 0.01, and the parameter ‘confidence’ was set to 0.1. After searching for possible rules, redundant rules were removed (see the supplement for the R code used in association mining).
We used Fisher’s exact test to determine whether to accept these rules, but the Bonferroni adjustment was not adopted. The reason for this choice was discussed above. A contingency table was designed for the test: N1 was the sample number corresponding to the LHS of the implication of an association rule; N2 was the number of samples fitting the description of both the LHS and RHS (right-hand side) of the implication; N3 was the total number of samples from one region; and N4 was the sample number corresponding to the RHS of the implication. Then, the odds ratio was (N2/N1)/(N4/N3), just the ‘lift’ provided by the function apriori.
In addition, we must note two major considerations. First, certain rules could be identified in Xishuangbanna and not in Dehong. Common support in the function apriori was used in the two regions, but the samples from Xishuangbanna are considerably less than those from Dehong. Second, the rules identified by apriori could be replaced by another simpler rule. The simpler rule shared a common RHS with the apriori rule, while its LHS was comparatively simpler. The rationality of such replacements should be confirmed.
The first consideration, whether the significant rules counted equally in both regions, was addressed with additional tests. When a rule was found to be significant in Xishuangbanna, another Fisher’s exact test was performed in Dehong. If a rule was significant in both regions but not in Dehong, we concluded that the sample size difference has a negligible effect on our analysis and that the regionality of the rules was not misjudged.
The second consideration, replacement of the rule identified by apriori, was addressed with the conditional probabilities based on the simpler rule and the apriori rule.
The possible simpler rules were exhausted with set operations. The LHS of the implication for an identified rule is seen as a set, and each element is a property-value pair (“Property” is the column name of the data set). Thereafter, all possible simpler rules can be listed by enumerating all the nonempty and proper subsets of the LHS of the identified rules.
The rationality of the replacement of an apriori rule with a simpler rule was determined with the ratio between two conditional probabilities. The first conditional probability was for the apriori rule, defined as N2/N1. N1 was the sample number corresponding to the LHS from this rule, and N2 was the sample number corresponding to both the LHS and RHS. The second conditional probability was for the simpler rule, defined as N4/N3. Similarly, N3 and N4 were the sample numbers corresponding to the LHS and to both the LHS and RHS of the simpler rule, respectively. Naturally, the ratio between the two probabilities was computed with (N2/N1)/(/N4/N3). This ratio is equivalent to the odds ratio generated by the Fisher’s exact test. When the ratio departs from 1 significantly, the simpler rule is thought to be distinct from the identified rule, and the replacement is not rational.
After the analysis above, the association rules in Xishuangbanna and property “Sex” was thought to be essential. Thus, an additional permutation test was conducted. In the Xishuangbanna data, the “Sex” label was shuffled for 10000 times, the Fisher exact test was applied to the rules including “Sex”. For each rule, the proportion of p value less than 0.01, 0.03 and 0.05 were recorded.
References
Ishikawa, I., Maeda, K., Nakai, S. & Kawaguchi, Y. Gender difference in the mean age at the induction of hemodialysis in patients with autosomal dominant polycystic kidney disease. American Journal of Kidney Diseases. 35(6), 1072–1075 (2000).
Gretz, N. et al. Gender-dependent disease severity in autosomal polycystic kidney disease of rats. Kidney International. 48(2), 496–500 (1995).
Li, F. et al. A Novel Mutation in CACNA1S Gene Associated with Hypokalemic Periodic Paralysis Which has a Gender Difference in the Penetrance. Journal of Molecular Neuroscience. 46(2), 378–383 (2012).
Kyriakou, A. et al. Gender differences in the prevalence and severity of bone disease in thalassaemia. Pediatr Endocrinol Rev. 6(Suppl 1), 116 (2008).
Toumba, M. & Skordis, N. Osteoporosis Syndrome in Thalassaemia Major: An Overview. Journal of Osteoporosis. 2010(1), 537673 (2010).
Moussavi, F. et al. Optimal method for early detection of cardiac disorders in thalassemia major patients: magnetic resonance imaging or echocardiography? Blood Research. 49(3), 182–6 (2014).
Yu Q. et al. Sex-specific association of rs4746172 of VCL gene with hypertension in two Han populations from Southern China. Scientific Reports 5, https://doi.org/10.1038/srep15245 (2015).
Ober, C., Loisel, D. A. & Gilad, Y. Sex-specific genetic architecture of human disease. Nature Reviews Genetics. 9, 911–922, https://doi.org/10.1038/nrg2415 (2008).
Reisner, E. G., Kostyu, D. D., Phillips, G., Walker, C. & Dawson, D. V. Alloantibody responses in multiply transfused sickle cell patients. Tissue Antigens. 30(4), 161–6 (1987).
Dhawan, H. K. et al. Alloimmunization and autoimmunization in transfusion dependent thalassemia major patients: Study on 319 patients. Asian Journal of Transfusion Science. 8(2), 84 (2014).
Datta, S. S., Mukherjee, S., Talukder, B., Bhattacharya, P. & Mukherjee, K. Frequency of Red Cell Alloimmunization and Autoimmunization in Thalassemia Patients: A Report from Eastern India. Advances in Hematology. 2015(10), 610931 (2015).
Woldu, D. O. & Haile, Z. T. Gender Roles and Perceptions of Malaria Risk in Agricultural Communities of Mwea Division in Central Kenya. Women & Health. 55(2), 227–43 (2015).
Esan A.J., Omisakin C.T., Titilayo O.E., Fasakin K.A. Gender difference on Stress induced Malaria parasite infection and effect of anti-malaria drug on Stress index. 2(3), 42–6 (1992).
Khadanga, S. et al. Severe Falciparum Malaria—Difference in Mortality among Male and Nonpregnant Females. Journal of Clinical & Diagnostic Research Jcdr. 8(12), 01–4 (2014).
Mengistu G., Diro E. Treatment outcome of severe malaria in adults with emphasis on neurological manifestations at Gondar University Hospital north west Ethiopia. Ethiopian Journal of Health Development. 20(2) (2006).
Chong, S. S., Boehm, C. D., Cutting, G. R. & Higgs, D. R. Sequence characterization of the |[ndash]|THAIallele of a thalassemia and rapid detection using a single-tube multiplex-PCR assay. Genetics in Medicine. 2(2), 103–103 (2000).
Yang, J. The prevalence and distribution of malaria in Dehongzhou Prefecture in 1951 - 2001. China Tropical Medicine. 4(5), 756–8 (2004).
Fan, J. H., Li, H. B. & Zhu, J. Evaluation of measures in malaria control in Xishuangbanna Prefecture from 2002 - 2006. China Tropical Medicne. 7(12), 2289–90 (2007).
Lin C.S. et al. http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CJFQ&dbname=CJFD7984&filename=KMYX1983Z1020&v=MDU3MDRkckt4RnRLbXJvOUhaSVI4ZVgxTHV4WVM3RGgxVDNxVHJXTTFGckNVUkwyZmIrZHBGeXJoVTd6TUxpRFM (1983)
Zhang, H. W. et al. Primary screening of hematology and analysis of thalassemia in Yunnan. Maternal & Child Health Care of China. 31(11), 2344–8 (2016).
Yang, Y. Q., Ge, S. J. & Fan, Y. H. Gene diagnostication and screening with regard to thalassemia of society colony in Yunan Dehong region(4 caes of family attached). International Journal of Laboratory Medicine. 31(6), 536–7 (2010).
Yao, L. Q. et al. A survey on anemia and thalassemia among minority children in Dehong Prefecture, Yunnan Province. Chinese Journal of Reproductive Health. 22(5), 1747 (2011).
Yao, L. Q. et al. Epidemiological study of thalasaemia among children in Xishuangbanna, Dehong and Nujiang of Yunnan province. Chinese. Journal of Medical Genetics. 28(5), 579–82 (2011).
Yao, L. Q. et al. Epidemiological Study on Thalassaemia Among the Children Under 7 Years Old From Yunnan Province With Unique Ethnic Minority Origin. Chinese. Journal of Obstetrics & Gynecology & Pediatrics. 9(3), 291–7 (2013).
Gan Z. J. http://kns.cnki.net/KCMS/detail/detail.aspx?dbcode=CJFQ&dbname=CJFD8589&filename=YXYZ198810010&v=MjgzNDBlWDFMdXhZUzdEaDFUM3FUcldNMUZyQ1VSTDJmYitkcEZ5cmhVTC9LUERYU2RMS3hGdG5OcjQ5RVpJUjg (1988).
Zhang, J. et al. Gene Mutation Spectrum of β-Thalassemia in Dai Ethinic Population of Two Border Region in Chinese Yunnan Province. Journal of Experimental Hematology. 24(1), 157–61 (2016).
Yao, L. Q. et al. To investigation on anemia among the children of Dai in different regions of Yunnan. Chinese Journal of Birth Health & Heredity. 2013(5), 136–9 (2013).
He, J. et al. Next-generation sequencing improves thalassemia carrier screening among premarital adults in a high prevalence population: the Dai nationality, China. Genetics in Medicine. (2017).
Li, Y. Y., Li, H. B. & Zhu, J. Epidemiology of malaria and control evaluation in Xishuangbanna prefecture, Yunnan, 1981- 2010. Disease Surveillance. 27(4), 280–1 (2012).
Qiu, Q. W. et al. Evidence of recent natural selection on the Southeast Asian deletion (–(SEA)) causing α-thalassemia in South China. Bmc Evolutionary Biology. 13(1), 1–10 (2013).
Elguero, E. et al. Malaria continues to select for sickle cell trait in Central Africa. Proceedings of the National Academy of Sciences of the United States of America. 112(22), 7051–7054 (2015).
Acknowledgements
This project was supported by grants from the National Science Foundation of China (Nos. 31500940 and 81260415), the Natural Science Foundation of Guangdong Province, China (No. 2016A030310037), the Shenzhen Peacock Program scientific research start-up foundation, and the National Science Foundation of Shenzhen University, China (No. C110205). This project was also supported by the foundation of the Natural Science Foundation of Yunnan Province, China (Nos. 2013DA008, 2014FC003, 2015DA008, 2014HB053, 2016YFC0900503, 2016RA037 and 2016RA038). We are grateful for help from the China National GeneBank, BGI-Shenzhen.
Author information
Authors and Affiliations
Contributions
Shenmin Guan, Yingying Zhao and Zhuo Xiao wrote the manuscript. Xin Song, Xiaorui Geng and Le Cheng reviewed the manuscript. Shenmin Guan conceptualized the research, planned the data mining, wrote all R codes, interpreted the previous study, and discussed all possible explanations, including malaria and gender-dependent symptoms. Xiao Zhuo provided Figs 1, 3 and 4 and edited Fig. 2. Wenhui Song cleaned the data and discussed the explanation relevant to malaria. Le Cheng and Xinhua Wu acquired the data and discussed gender-dependent symptoms. Yingying Zhao, Le Cheng, Xin Song, Jinlong Yang, Huanming Yang, and Jian Wang acquired the funding for this work.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guan, S., Zhao, Y., Zhuo, X. et al. Regional gender differences in an autosomal disease result in corresponding diversity differences. Sci Rep 9, 5472 (2019). https://doi.org/10.1038/s41598-019-41905-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-41905-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.