Abstract
The yield of pepper plants (Capsicum spp.) is their most important trait and is affected by the flower number and flowering time. Capsicum annuum produces a single flower per node and has an early flowering habit. By contrast, Capsicum chinense yields multiple flowers per node and has a late flowering character. However, the genetic mechanism underlying the control of these floral traits remains largely unknown. In this study, 150 F2 populations from an interspecific cross between the inbred lines 740 (C. chinense) and CA1 (C. annuum) and their parents were used to construct a molecular genetic linkage map using the specific length amplified fragment sequencing (SLAF-seq) technique. This linkage map, spanning 1,586.78 cM in length, contained 9,038 markers on 12 chromosomes, with a mean marker distance of 0.18 cM. Phenotypic data on the flowering time and flower number per node were collected over multiple years, and QTL analysis identified 6 QTLs for the flowering time and flower number per node by composite interval mapping (CIM) and genome-wide composite interval mapping (GCIM) methods at least in two environments. The candidate genes within the major QTL were predicted. In the major flowering time QTL, the candidate gene Capana02g000700, which encodes the homeotic protein APETALA2, was identified. Quantitative reverse-transcription PCR (qRT-PCR) analysis indicated that its expression level in 740 was higher than that in CA1. Gene expression analysis indicated that the expression of Capana02g000700 was significantly upregulated in flowers, and many floral development-related genes were found to be coexpressed with Capana02g000700, supporting the function of this gene in association with flowering time in C. chinense and C. annuum species.
Similar content being viewed by others
Introduction
Pepper (Capsicum spp.) is an economically important plant of the Solanaceae family, whose fruits are consumed as vegetables and food additives for their unique color, pungency, and aroma in many regions of the world, particularly in Asia and South and Central America1,2. Peppers are also grown for use as an ornament, for chemical industries, and for their pain-killing and medicinal properties3. Approximately 35 species of Capsicum are native to Central America4, of which Capsicum annuum, Capsicum chinense, Capsicum frutescens, Capsicum baccatum, and Capsicum pubescens have been domesticated and are now cultivated in different parts of the world5. Of these species, C. annuum is the one most widely grown, and its yield accounts of for 80% of pepper fruit production worldwide6. However, it is low in disease resistance and adaption to the humid lowland tropics, where, at least in Latin America, it has been replaced by C. chinense and C. frutescens6. Among the five domesticated Capsicum species, enormous variation were observed in plant architecture, flower-, leaf-, fruit-, metabolism- and disease resistance- related traits7,8. The yield of pepper is the most important trait, affected by factors such as the flower number and flowering time. Previous studies have strongly indicated that selection for more flowers and moderately early flowering can substantially enhance yields in various horticultural types of pepper6. C. annuum has one flower per node and early maturity, whereas C. chinense plants have multiple flowers (always two to four) per node and a late flowering time habit. Therefore, interspecific hybridization of C. annuum with C. chinense, with the multiple flower trait being transferred into C. annuum, may be potentially useful to increase the yield and enhance uniform maturity, which may make mechanical harvesting feasible9,10. However, transferring superior traits from C. chinense into C. annuum to develop viable commercial varieties is very time-consuming and expensive because of the number of backcrosses required. Since most of the traits mentioned above are quantitatively inherited or controlled by multiple major genes1,9,11, the discovery of the QTLs or major genes that govern these traits in various backgrounds is imperative; furthermore, the use of molecular-assisted selection can shorten breeding cycle and accelerate the breeding process of new varieties of pepper. During the past few decades, many genetic maps, including integrated maps, have been constructed for peppers using either intraspecific12,13 or interspecific8,14,15 populations to identify the QTLs of horticultural traits16. However, most of these genetic maps were low-density, and many identified QTLs covering large region lead hard to use for molecular assistant selection.
A genetic map, especially a high-density genetic map, provides an important foundation for QTL mapping and major QTL cloning. In Capsicum species, a number of interspecific and intraspecific genetic maps have been constructed by using conventional methods in previous studies1,15. However, the current number of markers is too small to build a high-density genetic map, which limits the efficiency and accuracy of QTL mapping. Next-generation sequencing (NGS) technologies can be used to detect large quantities of SNP markers in the whole genome for high-resolution genetic map construction. Several methods combine NGS with restriction enzyme digestion to reduce the complexity of the target genomes, including genotyping-by-sequencing (GBS-seq)17 and restriction site-associated DNA sequencing (RAD-seq)18. The selection of digested DNA fragment sizes is critically important to improve the efficiency of tag utilization. Unlike GBS-seq, which does not select the size of the digested fragment before PCR amplification, the RAD-seq conducts the size-selection step of the digested fragment before PCR amplification19. However, traditional RAD-seq technology has several shortcomings, such as more operation steps and shorter read length. By combining bioinformatics and RAD-seq technology, specific-locus amplified fragment sequencing (SLAF-seq) is developed20. SLAF-seq applies a bioinformatics approach to simulate the results of enzyme digestion, selects the most suitable restriction enzymes for double digestion, and then sequences the PCR-amplified fragments on an Illumina sequencer. This approach can effectively avoid repetitive sequences in the genome, develop SNP markers with uniform distribution in the genome, increase the effective reads obtained by sequencing, and improve the efficiency of molecular marker development20.
To date, many pepper intraspecific high-density genetic maps have been reported21,22,23. However, interspecific high-resolution genetic maps have rarely been reported in peppers24. Due to an abundance of polymorphic DNA sequences in interspecific individuals, the construction of a high-density genetic map based on SNPs markers of Capsicum species is possible. In this study, based on SLAF-seq, we constructed a high-density genetic map of an interspecific cross of C. annuum and C. chinense. Furthermore, we used a high-density genetic linkage map to detect QTLs for certain traits of peppers: flowering time and flower number per node.
Results
SLAF sequencing and genotyping of the interspecific cross F2 population
In this study, the 150 interspecific cross F2 populations and their parents were genotyped using SLAF-seq technology. Based on the results of the SLAF pilot experiment, HaeIII was used for SLAF library construction. The library comprised SLAF fragments ranging from 414 to 514 bp in length. After high-throughput sequencing of the SLAF library, 183.83 Gb of raw data was generated. In total, 749.26 M pair-end reads were obtained for both parents and 150 progenies, with an average of 4.92 M reads for each individual line. For quality control processing, reads of low-quality were discarded during quality checks in each cycle. This dynamic process was repeated until the average genotype quality score of all SLAFs reached the cut-off value of 30 (quality scores of at least 30, indicating a 1% likelihood of an error and thus 99.99% confidence). On average, Q30 was 92.78%, and the GC content was 38.21%. After reads clustering, 406,563 high-quality SLAFs were detected. The average depths of these SLAFs were 90.34 (male parent) and 58.61 (female parent) for parents and 11.74 for each individual progeny (Table 1).
Based on the results of SLAF positioning on the Zunla-1 (Capsicum annuum) genome, the SLAFs on each chromosome were calculated (Table 2), and a distribution diagram of SLAFs on each chromosome is shown in Fig. 1a,b. The SLAFs were distributed equally on each chromosome, and the pepper genome has been successfully simplified. Among the 406,563 high-quality SLAFs, 171,413 were polymorphic according to an analysis of allele numbers and the differences between gene sequences, with a polymorphism rate 42.16%. Of the 171,413 polymorphic SLAFs, 94,733 were classified into eight segregation patterns (ab × cd, ef × eg, hk × hk, lm × ll, nn × np, aa × bb, ab × cc, and cc × ab) (Fig. 1c). Because the F2 population was obtained from a cross of two diverse pepper inbred lines with the genotype aa or bb, only the SLAF markers that had segregation patterns of aa × bb were used in map construction.
SLAF polymorphism analysis. (a) All SLAFs (black lines) distributed on 12 chromosomes. (b) Polymorphic markers distributed on 12 chromosomes. (c) Number of markers for eight segregation patterns. The x-axis indicates eight segregation patterns of polymorphic SLAF markers; the y-axis indicates the number of markers. F2 population is obtained from a cross of two pepper inbred lines with the genotype aa or bb; therefore, only the SLAF markers, which had segregation patterns of aa × bb, were used in map construction.
High-density genetic map constructed with SLAF markers
To ensure the accuracy of genotyping, the following steps were performed as described during processing: (1) SLAFs with depth of less than 18-fold in each parent were discarded, (2) SLAFs with more than five SNPs were removed, (3) SLAFs with distorted segregation ratios (χ2 test, p < 0.05) were also maintained for genetic map construction, and (4) the markers demonstrating less than 85% integrity were discarded. Finally, 13,472 high-quality makers were distributed into 12 chromosomes according to their physical locations on the pepper reference genome and the MLOD scores >3 with other markers. As a result, 9,038 markers were designated for use in the final linkage map construction (Supplementary Tables 1 and 2). These markers are homozygous in the two parents, with a sequence depth 150-fold for the male parent and 101.4-fold for the female parent, respectively, and have more than 99% integrity of SLAFs for individuals. The polymorphic SLAFs used for the map construction ranged from 0.73% to 11.50% among the 12 linkage groups (LGs) (Table 2). Finally, the map contained 12 LGs and spanned a genetic distance of 1,586.78 cM in total, with an average distance of 0.18 cM between adjacent markers (Table 2). The distribution of the SLAF markers on each LG is shown in Fig. 2. On average, each LG contained 753 markers that covered an average of 132.23 cM. As shown in Table 2, the largest LG was Chr2, and it spanned a length of 188.3 cM, with 1,730 markers and an average distance of 0.11 cM between adjacent markers. In contrast, the smallest LG was Chr6, which harbored 1,355 makers, covered a length of 75.14 cM, and had a 0.06 cM average intermarker distance. The largest gap of 12 LGs ranged from 1.6 (Chr2) to 17.2 cM (Chr5). This genetic map included 28,741 SNPs (Table 2).
Evaluation of the Capsicum Genetic Map
The quality of the genetic map was evaluated by using HighMap to construct haplotype map and heat map. The haplotype maps, which reflect the double recombination and deletion of the population, were generated for the parental controls and 150 offspring using 9,038 SLAF markers. In this study, most of the recombination blocks were distinctly defined. The missing data for each LG ranged from 0.76% to 1.14% (Supplementary Table 3). Most of the LGs were uniformly distributed, suggesting that the genetic maps were of high quality. The heat maps showed the relationships of the recombination between markers from each LG. The comparisons between markers were used to assign recombination scores to the 9,038 SLAF markers, after which the heat maps were constructed. The resulting maps showed that the order of the SLAF markers in most of the LGs have been correctly ordered (Supplementary Fig. S1). In total, 803 distorted SLAF markers were integrated into the map (Supplementary Table 4). They were noted in all LGs except Chr2 and Chr3, and most of the distorted markers were skewed toward the male parent. The frequencies of the distorted markers in Chr4 and Chr12 were much higher than those of the other LGs.
To evaluate the collinearity between the genetic map and the Zunla-1 reference genome, all SLAF markers were anchored on the Capsicum reference genome. As the results presented in Fig. 3, a sufficient genome coverage and the accurate genetic location of the markers was revealed by the consecutive curves except for Chr1 and Chr8. Nevertheless, the Spearman rank correlation test of the genetic map and the physical map revealed that the correlations were significant (p < 0.001) among the 12 chromosomes, indicating that the 9,038 SLAF markers were accurately positioned on 12 chromosomes, and the Capsicum genome was sufficiently covered with these SLAF markers. The genetic arrangements of most markers were also considered to coincide with their physical direction based on the falling trend of the curve.
Collinearity of 12 chromosomes with the Capsicum reference genome. The x-axis indicates the genetic distance of the Capsicum chromosomes, and the y-axis represents the linear order of the physical position in the Capsicum genome. All 9038 SLAF markers in these chromosomes are plotted as dots on the figure. Different colors indicate different chromosomes. Spearman’s correlation coefficient of the genetic map and the physical map was presented in the bottom of the figure.
QTL analysis using the high density genetic map
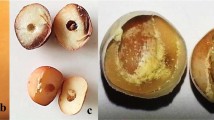
For floral phenotypic data collection, populations were planted in a greenhouse. As shown in Fig. 4, flowering time and flower number per node from the inbred line of 740 and CA1 displayed obviously different habits. For the inbred line of 740, the plants were with late flowering (flowering-score was 1), with multiple flower number per node (Fig. 4a,c; Supplementary Table 5 and 6). In contrast, the inbred line of the CA1 plants were early flowering (flowering-score was 6), with one flower per node (Fig. 4b,d; Supplementary Tables 5 and 6). As a result, the fruit in the parental and offspring populations mainly ranged from 1 to 3 per node (Fig. 4e–h).
Phenotypes of the inbred lines 740 and CA1 and their F1 and F2 populations. (a,b) The 740 and CA1 plants in the flowering stage. (c,d) A node close-up of the 740 and CA1 plants, with the 740 plants showing multiple flowers per node and the CA1 plants showing one flower per node. (e,f) 740 and F1 individual fruits in the mature stage. (g,h) Pepper F2 individuals with two fruits (g) Or one fruit (h) Per branch node, respectively.
Phenotypic data of flowering time and flower number per node of the two parents, and the F1 and F2:3 families were collected from three environments across three years. In all experiments, the phenotypic data flowering time of the two parents (740 and CA1) were located at the extreme ends of the largely normally distributed family means, and those for the F1 and F2:3 families were close to the mid-parent values, suggesting the quantitative genetics of these traits (Supplementary Fig. S2; Supplementary Tables 5 and 6). For the multiple flower trait, the CA1 parent averaged only one flower per node compared to 2.3 for 740. The F1 produced approximately 1.7 flowers per node, whereas the F2:3 were observed with the mean (1.5 flowers/node) being skewed toward the CA parent, suggesting incomplete dominance of the character. The coefficients of variation (CV%) of flowering time (47.79–50.01%) and multiple flowers (34.87–36.24%) in the F2:3 populations were significantly higher than the corresponding CV% from parents and F1, indicating the existence of real variation in our populations. Despite being collected over different years, data for these traits were highly consistent and of good quality, which provided a solid foundation for the QTL analysis.
QTL analysis of flowering time and flower number per node traits
We conducted QTL analysis using the composite interval mapping (CIM) approach with data for each year in R/QTL. The LOD threshold for declaring significance of a QTL for flowering time and flower number per node was determined with 1000 permutations (p = 0.05). Details of each detected QTL, including the map location, LOD value, percentages of total phenotypic variances explained (R2), and 1.5-LOD support interval are presented in Table 3 and Supplementary Table 7, and the related SLAFs sequence are listed in Supplementary Table 1. As shown from the results, three flowering time QTLs, Ft2.1, Ft6.1 and Ft6.2, and three flower number per node QTLs, Mf2.1, Mf7.1 and Mf10.1, were consistently detected in all three environments. These QTLs could explain 46.35–50.37% and 99.13–99.76% of flowering time and flower number per node of the phenotypic variations, respectively. For the three flowering time QTLs, Ft2.1 had the largest effect, which accounted for 24.59–27.87% of the phenotypic variations. With regard to the three flower number per node QTLs, Mf2.1 had the largest effect, explaining approximately 40% of the phenotypic variations, and this was followed by Mf7.1 and Mf10.1, which explained approximately 30% of the phenotypic variations. Of these QTLs for the detected traits, the interval ranged from 0.382 cM to 5.343 cM, and the peak LOD score ranged from 2.45 to 7.59. Combining the results from the QTL analysis with the QTLs for the three flowering times and the three flowers per node average explained 48.62% and 99.43% of the phenotypic variations across the three years, respectively. To obtain a more reliable QTL analysis, genome-wide composite interval mapping (GCIM) was performed with the QTL.gCIMapping.GUI package under a random model. QTLs were identified on Chr2, Chr 6, Chr7, Chr8, Chr10 and Chr11. For the flowering time traits, the QTLs detected with the CIM method were also identified by the GCIM method at least in two environments. The GCIM detected QTL peak LOD scores ranging from 5.16 to 22.7, which were higher than the QTL peak LOD score ranging from 2.45 to 5.43 and detected by the CIM method, and the detected QTL interval with the GCIM method was narrower than the corresponding QTL identified by the CIM method. The results for these QTLs explained 4.96–33.5% of the observed phenotypic variation phenotypic variations. For the flower number per node, the QTLs detected by the CIM method were consistently detected by the GCIM method. The QTLs peak LOD score detected by the GCIM were higher than the corresponding QTLs detected by the CIM, while the QTLs explained the observed phenotypic variation phenotypic variations were decreased. Nevertheless, two novel minor QTLs (i.e., Mf8.1, Mf11.1) were also detected by the GCIM method in at least two environments. The results for these QTLs explained 5.84–38.16% of the observed phenotypic variation phenotypic variations. These QTLs could explain 29.99–60.62% and 72.47–82.52% of flowering time and flower number per node of the phenotypic variations, respectively.
Prediction of gene control flowering time and multiple flower trait
To test the accuracy and precision of our genetic map, given that both the flower time Ft2.1 and flower number per node Mf2.1 QTL are the most reliable and can explain most phenotypic variation among the detected QTLs, the QTLs underlying the control of flowering time Ft2.1 and flower number per node Mf2.1 were used for gene prediction. For the Ft2.1, QTL region was mapped to 0.763 cM, and the interval physically represents approximately 210 kb in the Zunla-1 reference genome, with 25 putative predicted genes being included (Fig. 5a; Supplementary Table 8). Strikingly, Capana02g000700 (85140186bp-85144310bp on chromosome 2), which encodes a floral homeotic protein, APETALA 2, was identified. Phylogenetic analysis indicated that the Capana02g000700 homologs from Antirrhinum LIP1 and LIP225,26, Arabidopsis thaliana APETALA227, and pepper CaAP228 play an important role in floral development (Fig. 5b). Analysis of the nucleotide sequence of Capana02g000700, 8 SNPs and a 6 bp deletion were detected among the 740 and CA1 ORF regions (Supplementary Fig. S3), which lead to predicting that the protein from CA1 has two amino acid deletions and five amino acid changes, including an amino acid in the AP2 domain (Supplementary Fig. S4). The qRT-PCR analysis of Capana02g000700 expression indicated the expression was significantly more upregulated in the floral than other detected tissues (Fig. 5c). A comparison of the 740 and CA1 mRNA amount in the different tissues indicates its expression in 740 was significantly higher than in the corresponding CA1 (Fig. 5c). The RNA-Seq expression data in flower, leaf and fruit differential developmental stages were retrieved from the pepper inbred line 6421 (C. annuum)29, and Capana02g000700 was found to have a higher expression level in pepper flower than in other tissues (Fig. 5d), which indicates that it plays an important role in flower development, strongly suggesting that Capana02g000700 is candidate gene for transcriptional repression in the control of flowering time of pepper.
Local genetic linkage map showing the major QTL and identification of a floral homeotic protein, APETALA2, located in the major QTL region. (a) The major QTL for flowering time mapped to the interval between markers Marker6530388 and Marker6433087 on chromosome 2. The black arrow indicates the predicted genes in the interval, and the red arrow indicates the candidate gene Capana02g000700, which is associated with flowering time. (b) Phylogenetic tree of Capana02g000700 and its homologs. Numbers indicate the percentage of bootstrap support for each branch (1000 replicates). PhROB3 (APG29274.1), SlAP2b (NP_001233908.2), AmLIP1 (AAO52746.1), AmLIP2 (AAO52747.1), CsAP2 (AFK29251.1), QsAP2 (XP_023883493.1), AtTOE3 (NP_201519.1), AtTOE2 (NP_001189625.1), AtAPETALA2 (NP_195410.1), AtFOL2 (OAO99308.1), and CaAP2 (AJC11181.1). (c) qRT-PCR analysis of Capana02g000700 expression in different 740 and CA1 tissues. Data presented are mean values of three biological repeats with three biological replicates s.d. **P < 0.01 (Student’s t-test). (d) Digital gene expression level of Capana02g000700 presented as FPKM values in the inbred line 6421 (C. annuum) from different tissues at different developmental stages. AL, all leaves; AR, all root; AS, all stem; P10, fully blossomed flower petals; O10, ovary with stigma; STA10, stamen.
With regard to the Mf2.1, the QTL region was mapped to 0.382 cM, and the interval physically represents approximately 1400 kb in the Zunla-1 reference genome, and 98 putative predicted genes were found to reside in this region (Supplementary Fig. S5a; Supplementary Table 9). Inflorescence architecture is based on changes in the activity of the meristems, small groups of stem cells located at the tips of shoots30,31. Studies illustrated that, during the vegetative transition to flowering, the dynamically expressed genes were found enriched for transcription factors32. In the Mf2.1 region, GO and COG analysis indicate that the genes involved in the transcription regulation process were enriched (Supplementary Fig. S5b). We retrieved the pepper transcriptome data of the middle vegetative meristem (MVM), transition meristem (TM), and floral meristem (FM) tissues reported previously32. Expression analysis revealed that a large number of genes were upregulated in TM, including TF from different family such as GATA (Capana02g002708, Capana02g002714), NAC (Capana02g002682), Zinc-finger homeodomain protein (Capana02g002717), and bHLH (Capana02g002736) (Supplementary Fig. S5c). These upregulated TFs within the Mf2.1 region may be the candidates involved in control of transition meristem to form of floral meristem and then determine flower numbers.
Coexpression analysis of genes expression
We adopt the WGCNA to identify genes with differential expression at distinct stages of flower development. We identified a flower-development-specific module, which contained 107 genes (Fig. 6a; Supplementary Table 10). Of these genes, ten transcription factors (TFs), including Capana02g000700, were identified, indicating that these TFs may be involved in the transcriptional control of flower development. Strikingly, among these genes, nine were found highly co-expressed with Capana02g000700, and three TF Capana11g000298 (bZIP family), Capana08g000623 (MADS-box family), Capana05g001110 (bHLH family) were detected as coming from different families (Fig. 6a). Capana08g000623, an orthologous gene from A. thaliana PISTILLATA, was reported as being involved in flower development and its expression regulated by the AtAPETALA233. In tobacco plants, reduced amounts of TGA2.1 (Capana11g000298 orthologous gene) from tobacco resulted in the development of petal-like stamens, indicating a regulatory role of TGA2.1 in defining organ identity in tobacco flowers34. In addition, Capana05g001110 encoding brassinosteroid enhanced expression 3 (BEE3) homologue positively regulated brassinosteroid signaling and required flower normal growth in Arabidopsis35. A heat map shows the expression (FPKM) of 65 genes selected from Fig. 6a and found most of the expression of these genes was flower-development regulated (Fig. 6b). These results strongly support that Capana02g000700 may repress target genes and/or TFs expression to regulate flower development and then affect flowering time.
Co-expression analysis genes associated with flower development in pepper. (a) Co-expression analysis of genes co-expressed with Capana02g000700. The light green colour indicates the genes that are most highly co-expressed with Capana02g000700. (b) Heat map displaying 65 expressed genes selected from (a) pepper in differential development stages. AL, all leaves; AR, all roost; AS, all stems; P10, fully blossomed flower petals; O10, ovary with stigma; STA10, stamen.
Discussion
High-density genetic map constructed with C. chinense and C. annuum interspecies hybridization F2 population
The pepper genome sequence has been completed, and the size of the pepper genome is large, estimated to be 3.48 Gb3,36. Whole-genome deep resequencing or low-coverage sequencing is relatively costly for large genomes and usually unnecessary for gene/QTL mapping11,37. The SLAF-seq, which was developed based on high-throughput sequencing, is an effective strategy for large-scale SNP discovery and genotyping20. In contrast to conventional methods, which are inefficient, expensive, and time-consuming20,38, SLAF sequencing can generate large amounts of sequence information and handle whole genome density distributions, which ensures density, uniformity, and efficiency of marker development20,39,40. In this study, we constructed a high-density genetic map using the SLAF-seq technology with an interspecies F2 population consisting of 150 individuals. This map, which included 9,038 high-quality SLAF markers on 12 LGs, covered a genetic distance of 1,586.78 cM in total, with an average distance of 0.18 cM between adjacent markers, and showed 99% integrity for individuals (Fig. 3, Table 2). To date, compared with other genetic maps of Capsicum species16,24,41,42, the genetic map reported in this study is the highest density map and had the smallest average distance between interval markers for Capsicum genus plants. With this high-density map combination with high-quality genome sequences, the candidate genes within a narrow region interval between adjacent markers can be predicted directly. However, as reflect in our results we also need to keep in mind that success QTL mapping depend on the chromosome region the QTL falls into. Overall, SLAF-seq technology is ideal for population genotyping and for high-resolution linkage map construction because of its high success rates, specificity, and stability20. Accordingly, the genetic map could be used for detecting QTL for important horticultural traits, and the narrowed QTLs also provided several promising candidate genes for further functional identification.
Segregation distortion is a commonly observed in both interspecific and intraspecific cross populations43. In this study, to avoid the losing information, the 803 SLAF markers that were also utilized in map construction. In total, almost all the segregation makers were skewed toward the male parent. The distorted segregation could be caused by gametophytic factors that affect female gametes43,44, but this distortion needs further study. We found that the rate of the polymorphic SLAFs in the Chr1 and Chr8 being used for map construction were significantly lower than other LGs. Possibly, chromosome translocation between chromosome 1 and chromosome 8 in the inbred pepper lines 740 and CA1 and the many markers had to be filtered before being used for genetic map construction. Indeed, the translocation between chromosome 1 and chromosome 8 in C. frutescens, C. chinense, C. baccatum and wild C. annuum compared to cultivated C. annuum was previously reported and has been well characterized45. In addition, only C. annuum-originated sequences were used for developing the SLAF markers, whereas some C. chinense specific markers were unable to be detected. Therefore, the alignment of the SLAFs to the C. chinense genome needs to be developed for a more complete genetic map in the future.
QTL identification of floral-related traits
The yield-related traits are important for pepper production, and high yield and high disease resistance always are the most important objectives for pepper breeding. Therefore, the detection of QTLs or genes for these traits should be important for pepper genetic improvement. Hitherto, QTLs studies on yield related traits have been widely documented in intraspecific and interspecific crosses of Capsicum species populations8,12,46,47. Previous studies have strongly indicated that selection for early flowering can enhance yields in various horticultural types of pepper. Flowering time is a fully quantitative complex trait, and researchers always record the number of days between sowing and anthesis or at certain day after sowing to score the flower or fruit developmental status of the third node to evaluate flowering times8,15. In addition, some others also used the leaves numbers on primary stem to evaluate flowering time28. After comprehensively comparing the characteristic of parents and progenies used in this and previous studies, we selected the score criterion for evaluating the flowering time, and three QTLs were detected in the study (Table 3). For flowering time, the major QTL Ft2.1 explains approximately 30% of the phenotypic variation detected on Chr2 in our populations across the three years. The major QTL related to flowering time in the Chr2 was also detected by using the RILs derived from the interspecific cross of the C. frutescens × C. annuum15. A comparison of the physical position reported previously, adjacent to the Ft2.1 QTL interval, revealed a gene previously reported and mapped on the Chr2 from different studies14,28, which promotes the phase transition from inflorescence meristem to floral specification in pepper CaAP2 (Capana02g003062). In addition, in the C. annuum intraspecific F2 populations, using an SLAF-seq and BSA analysis, a candidate region was mapped on Chr12 that controls the first flower node and determines the flowering time47. However, the Ft2.1 not located in a previously reported physical position, indicated the Ft2.1 is a new locus that plays an important function in controlling flower development in the Capsicum genus among plants of different genetic backgrounds. In addition, among the two QTLs detected on Chr6, the novel QTLs Ft6.1 and Ft6.2 detected in this study differ from previous studies, suggesting that perhaps the population used for QTL identification came from different species, as previously reported. However, the three detected QTLs, which accounted for approximately of 50% of the total phenotypic variance, were detected with two methods, which may have been because the flowering time was scored based on a visual scale of 1–6, which limits the resolution of mapping a fully quantitative trait. Alternatively, markers with significant segregation distortion were used for genetic map construction, and these distorted markers may have affected the QTL analysis43,44,48. A more detailed scoring criterion may contribute to detecting more QTLs for the flowering time.
Possibly, the gene for the multiple flower trait could be transferred from the C. chinense to C. annuum varieties with a more concentrated fruit set, thereby contributing to a potentially higher yield10. In this study, we found the hybrid progenies with multiple flowers plants can produce an average of approximately 1.5 flowers per node (Supplementary Table 6). In addition, we observed that progeny with multiple flowers are always accompanied with more than one fruit setting in a node (Fig. 4), and this result was consistent with results reported previously by others9,10. Previous studies have proposed hypotheses for different genetic control mechanisms of multiple flowers: (i) a three-gene dominance model with epistasis49, (ii) at least five independently segregating chromosome segments involved in the multiple-flower habit9, (iii) seven semi-recessive additive genes from C. chinense50, and (iv) three major dominant genes from C. chinense10. Possibly, the populations were delivered from differential accessions of C. annuum and C. chinense, and the interrelationships among these models are unclear. In this study, with the CIM and GCIM QTL analysis, the detected QTLs Mf2.1, Mf7.1 and Mf10.1, with a positive additive effect (increased flower number), could exhibit greater phenotypic variation of the multiple flower trait. However, the GCIM method was also able to detect another two QTLs Mf8.1 and Mf11.1. In addition, the QTLs detected by GCIM had higher LOD peak scores, had more narrowed interval and were greater in number, indicating that GCIM mapping approach is more effective and reliable for detect more QTLs. Over all, the result seems consistent with the hypothesis of five-gene model of genetic control of multiple flowers in Capsicum9,10, but this needs further study. Nevertheless, we provided more detailed loci information, and this could contribute to uncovering the underlying genetic and molecular mechanism of multiple flowers traits in C. chinense. In addition, three QTLs related to multiple flowers Mf2.1, Mf7.1, Mf8.1, Mf10.1 and Mf11.1 were mapped in a narrow interval, which means that molecular markers closely linked to multiple-flower-per-node locus can be more effectively applied in early selection. Within the Mf2.1 region, even though we identified many TFs and found the expression of these TFs were upregulated in the transition meristem, these TFs may play an important role in control of vegetative meristem transition to floral meristem, which determines inflorescence architectures (flower number determinants). However, we found ANANTHA (AN) and COMPOUND INFLORESCENCE (S, the homolog of WUSCHEL-RELATED HOMEOBOX 9, WOX9) and their orthologs genes have conserved functions in the control of inflorescence architecture among Solanaceae species31,51. Given that the QTL can vary according to various factors such as mapping populations, genotyping method, detection method and environmental factor43,44,52. When considering these factors, we cannot rule out the possibility that these genes were involved in the control of the trait of flower number per node between C. annuum and C. chinense species. Further study is needed to finely map and identify the candidate genes underlying molecular mechanism control of multiple flowers and for the development of reliable makers for marker-assisted selection to pyramid the genes that control multiple-flower into commercial cultivars.
Multiple strategies to prediction of flowering time candidate genes
Within the flower major region Ft2.1, we were able to identify a candidate gene for the regulation of flowering time and flower development. After annotation of the 25 genes resides in genomic regions, we found Capana02g000700 encoding a floral APETALA2 protein, whose homologs from AtAPETALA2 and AtFLO2 act as flowering suppressors (Fig. 5b). In addition, we found Capana02g000700 expression significantly upregulated in flowers, and we also determined its expression in late flowering 740, where the expression was significantly higher than observed for the early flowering CA1 (Fig. 5c). Furthermore, we found many flower developmental-stage-specific genes were highly coexpressed with Capana02g000700 (Fig. 6). In a sequence comparison of CDS between the parents used for the QTL mapping, we detected 8 SNPs and a 6 bp deletion in CA1, but none of these changes result in a premature stop codon (Supplementary Figs S3, S4). We cannot rule out the possibility that the sequence variations between 740 and CA1 may have caused change in the gene activity. However, after comparing the Capana02g000700 gene from the C. annuum and C. chinense reference genomes reported before, we found the SNPs were highly conserved among the same species (Supplementary Figs S3 and S4), and we assumed that no major changes in the activity of the protein could be expected. In Arabidopsis, one of actions of AtAPETALA2 was to control flower development by repressing the flowering-promoter MADS-box transcription factor such as PISTILLATA33, AGAMOUS40 and SOC141. So, the results presented in this study show that the higher expression of Capana02g000700 in late flowering 740 results in enhanced repression of target flowering-promoter genes expression, which finally leads to late flowering. Although we could illustrate significant expression differences between the early flowering and late flowering parents, the underlying specific sequence variation in the promoter region associated with this difference is still unknown. We retrieved the C. chinense and C. annuum genome sequence reported previously3,36,53, and we found that the nucleotide sequence within the Ft2.1 region was misassembled or that the assembly quality was quite low for many of the gaps presented. We tried to elucidate the DNA variations between CA1 and 740, but we failed to amplify of Capana02g000700 promoter using the primers designed for the C. annuum genome sequence (Zunla-1 and CM334)3,36, indicating that a larger transposon may exist in its promoter. Because the gene detected in this study was different from the previously reported CaAP2, we assume the Capana02g000700 detected in this study is CaAP2 and Capana02g000700, which evolved from the same ancestor gene but evolved after the two AP2/ERFs (i.e., CaAP2 and Capana02g000700). These genes, which evolved independently, control flowering time in a different genetic background. We hypothesized that the Capana02g000700 mainly exerts control on the flowering time between C. annuum and C. chinense species, but further research is needed.
Materials and Methods
Plant materials
To develop a F2 population of interspecific hybridization between C. chinense and C. annuum, a cross was made between the inbred lines 740 (C. chinense) and CA1 (C. annuum). The female parent 740 plants multiple flowers per node and late flowering. In contrast, the plants of male parent CA1 has one flower per node and early flowering. The two parental lines were planted in greenhouse in the spring of 2013, the F1 of 740 × CA1 were grown in autumn of 2013 and self-pollinated to obtain 150 F2 and the F2 population were planted in the spring of 2014 and self-pollinated to obtain 150 F2:3 families.
Phenotypic data collection
Phenotypic data were collected in three environments over 3 years. The parents, F1 and F2:3 (2014 autumn, 2015, 2017 spring) families were grown in green house or College of Horticulture at Guangzhou, China. The 740, CA1, F1 and F2:3 families were designed with two replicate, and planting with 8 plants in each replication. Flowering time-scores of one to six were given based on the developmental stage of the flower/fruit at the third node on day 90, and the scoring of individuals was according to previous reported method with little modifications8. The scores represented the following traits: 1 = no obvious flower bud, 2 = flower bud/flower, 3 = small fruits, 4 = small- to medium-size fruits, 5 = medium-size fruits and 6 = mature fruits. With respected to multiple flowers traits, the survey was completed after the flower number at the fifth node could be clearly distinguished, and plants were evaluated the numbers of the flowers produced per node for the first five sets of nodes. We measured the traits on individual plants, and averaged within each F3 family.
DNA extraction
DNA was extracted from young health leaves of two parents and the 150 F2 offsprings by the method of CTAB54. The DNA samples were quantified with NanoDrop ND-1000 spectrophotometer (Wilmington, USA) and by electrophoresis in a 1% agarose gels with lambda DNA as a standard.
SLAF library construction and high-throughput sequencing
The SLAF-seq strategy was used to analyse the genotype of two parents and 150 F2 offspring as described previously20. Briefly, the genomic DNA of the two parents and 150 F2 populations was digested using the HaeIII restriction enzyme (New England Biolabs, USA). Subsequently, a signal nucleotide overhang was added to the digested fragments along with Klenow fragments (New England Biolabs, USA) and dATP at 37 °C. Then, PAGE-purified Duplex Tag-labelled sequencing adapters (Life Technologies, USA) were ligated to the A-tailed DNA with T4 DNA ligase (New England Biolabs, USA). After incubation, the reaction products were pooled and purified using a Quick Spin column (Qiagen, Germany). The purified products were electrophoresed on a 2% agarose gel, and fragments with sizes ranging from 414 to 514 bp were collected and purified using a gel extraction kit (QIAGEN, Germany). The purified product was sequenced on an Illumina HiSeq. 2500 system (Illumina, USA) according to the manufacturer’s instructions.
Sequence data analysis and genotyping
The SLAF marker grouping and genotyping were performed using procedures as described previous20. Briefly, after filtering out the low-quality reads (quality score < 30e), the remaining cleaned SLAF pair-end reads were clustered based on sequence similarity as alignment with BLAT (-tilesize = 10 -stepsize = 5). Subsequently, pair-end clean reads were mapped onto the reference genome of C. annuum. var Zunla-136 and reads with over 90% similarity sequences were grouped into one SLAF locus. Minor allele frequency evaluation was used to define alleles in each SLAF locus. C. annuum and C. chinense are diploid species, one locus could harbor at most four SLAF tags, locus containing more than four tags were filtered out as repetitive SLAFs, and those with two, three, and four tags were identified as polymorphic SLAFs. Then polymorphic SLAFs were classified into eight segregation patterns (aa × bb, ab × cc, ab × cd, cc × ab, ef × eg, hk × hk, lm × ll and nn × np). Because the F2 population is obtained from a cross of two diverse pepper inbred line with the genotype aa or bb, therefore only the SLAF markers which had segregation patterns of aa × bb were used in map construction.
Sequence data analysis and genotyping
The SLAF marker grouping and genotyping were performed using procedures as described previous20.
Briefly, after filtering out the low-quality reads (quality score < 30e), the remaining cleaned SLAF pair-end reads were clustered based on sequence similarity as alignment with BLAT (-tilesize = 10 -stepsize = 5). Subsequently, pair-end clean reads were mapped onto the reference genome of C. annuum. var Zunla-136 and reads with over 90% similarity sequences were grouped into one SLAF locus. Minor allele frequency evaluation was used to define alleles in each SLAF locus. C. annuum and C. chinense are diploid species, one locus could harbor at most four SLAF tags, locus containing more than four tags were filtered out as repetitive SLAFs, and those with two, three, and four tags were identified as polymorphic SLAFs. Then polymorphic SLAFs were classified into eight segregation patterns (aa × bb, ab × cc, ab × cd, cc × ab, ef × eg, hk × hk, lm × ll and nn × np). Because the F2 population is obtained from a cross of two diverse pepper inbred line with the genotype aa or bb, therefore only the SLAF markers which had segregation patterns of aa × bb were used in map construction.
Genetic map construction
In order to ensure the quality of genetic map, high-quality SLAF markers for the genetic map construction were filtered by the following criterions: (1) SLAF makers with parents sequence depth less than 18 were filtered out; (2) a SLAF which had less than five SNPs and average depth of each sample above four, was defined as a high quality SLAF marker; (3) markers with complete less than 85% were filtered; (4) Since distortedly segregated markers are ubiquitous and would affect the mapping construction and QTL analysis43,44,48, partial distorted polymorphism markers showing significance (p < 0.05) were maintained to construct the map. Subsequently, by using the HighMap strategy, the SLAF markers were assigned into chromosomes based on the pepper genome, and 12 linkage groups (LGs) were obtained. The modified logarithm of odds (MLOD) value was calculated between two adjacent makers, the SLAFs with MLOD values less than three were excluded. In addition, using HighMap software to analyze the linear array of markers in each LG, and estimate the genetic distances between two adjacent markers.
QTL/Gene mapping
Plant trait QTLs were identified by different methods. QTLs were detected by CIM methods with the R/QTL package methods using R/QTL v3.1.155. The significance thresholds were determined using 1,000 permutations (p < 0.05). The results from the CIM analysis were used to construct the QTLs, and their positions were used in a default model. In addition, multilocus QTL mapping was performed by the software of QTL.gCIMapping.GUI (https://cran.r-project.org/web/packages/QTL.gCIMapping/index.html) according to the user manual52,56.
Candidate gene selection and annotation
The predicted genes in the target QTL region were analysed according to the annotation of the pepper Zunla-1 and CM334 reference genomes3,31. The function of genes detected in the candidate region was manually confirmed using protein BLAST. In addition, GO enrichment and KEGG pathway analyses were performed with default settings. Multiple sequence alignments were performed with ClustalX, and a phylogenetic tree was calculated by the neighbour-joining method and bootstrap analysis with 1000 replicates via MEGA7 software57.
qRT-PCR analysis candidate gene expression level
Floral buds, fruit at developmental stage of 16 day post anthesis, leaves at 20 day after emergence, stem and root were collected from 740 and CA1, respectively. Samples were ground into fine powder in liquid nitrogen, and then the RNA was isolated from all samples using HiPure Plant RNA Mini Kit (Magen, China). Subsequently, the RNA from each sample was used for the reverse transcription reaction using a HiScript Q RT SuperMix for qPCR reagent kit with gDNA wiper (Vazyme, China). Quantitative real-time PCR analysis was performed on a LightCycler 480 Real-Time PCR System according to the manufacturer’s instructions; the qPCR program was according to described previously2 and primers used for analysis were listed in Supplementary Table 11. The reported values represent the mean of three biological replicates.
Candidate gene cloning and sequence analysis
When the candidate gene sequence was cloned, the total RNA was extracted from the bud of the CA1 and 740 by using a HiPure Plant RNA Mini Kit (Magen, China). The RNA from each sample was used for the reverse transcription reaction using an ImProm-II Reverse Transcription System (Promega, USA). The cDNA samples served as the template for amplification with the LA Taq DNA polymerase (TaKaRa, Japan) for the gene-specific marker (Supplementary Table 11), and the PCR products were cloned into a pMD19-T vector (TaKaRa, Japan). Positive clones were picked to culture for plasmid extraction and sequencing.
Co-expression analysis of gene expression
Co-expression network of gene expression was constructed with the weighted gene coexpression network analysis (WGCNA) package58 using gene expression data of different tissue samples from different developmental stages29. The modules were obtained using the automatic network construction function blockwiseModules with default settings. The genes co-expressed network was visualized by the Cytoscape 3.059.
Data Availability
Sequence data accession number (SUB4758068). Capana02g000700 sequence of inbred line CA1 accession number: 2165004; Capana02g000700 sequence of inbred line 740 Accession Number: 2165018.
References
Kang, B. et al. An interspecific (Capsicum annuum × C. chinese) F2 linkage map in pepper using RFLP and AFLP markers. Theor. Appl. Genet. 102, 531–539 (2001).
Zhu, Z. et al. Pyramiding of AtEDT1/HDG11 and Cry2Aa2 into pepper (Capsicum annuum L.) enhances drought tolerance and insect resistance without yield decrease. Plant Cell Tiss. Org. 120, 919–932 (2015).
Kim, S. et al. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet. 46, 270–278 (2014).
Carrizo Garcia, C. et al. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann. Bot. 118, 35–51 (2016).
Perry, L. et al. Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the Americas. Science 315, 986–988 (2007).
Pickersgill, B. Genetic resources and breeding of Capsicum spp. Euphytica 96, 129–133 (1997).
Lefebvre, V. et al. QTLs for resistance to powdery mildew in pepper under natural and artificial infections. Theor. Appl. Genet. 107, 661–666 (2003).
Rao, G. U., Ben Chaim, A., Borovsky, Y. & Paran, I. Mapping of yield-related QTLs in pepper in an interspecific cross of Capsicum annuum and C. frutescens. Theor. Appl. Genet. 106, 1457–1466 (2003).
Tanksley, S. D. & Iglesiasolivas, J. Inheritance and transfer of multiple-flower character from Capsicum-chinense into Capsicum-annuum. Euphytica 33, 769–777 (1984).
Shuh, D. M. & Fontenot, J. F. Gene-transfer of multiple flowers and pubescent leaf from Capsicum-chinense into Capsicum-cnnuum backgrounds. J. Am. Soc. Hortic. Sci. 115, 499–502 (1990).
Xu, X. et al. Mapping of a novel race specific resistance gene to phytophthora root rot of pepper (Capsicum annuum) using bulked segregant analysis combined with specific length amplified fragment sequencing strategy. PloS One 11, e0151401 (2016).
Chaim, A. B. et al. QTL mapping of fruit-related traits in pepper (Capsicum annuum). Theor. Appl. Genet. 102, 1016–1028 (2001).
Lu, F. H. et al. SNP marker integration and QTL analysis of 12 agronomic and morphological traits in F(8) RILs of pepper (Capsicum annuum L.). Mol. Cells 34, 25–34 (2012).
Tan, S. et al. Construction of an interspecific genetic map based on InDel and SSR for mapping the QTLs affecting the initiation of flower primordia in pepper (Capsicum spp.). PLoS One 10 (2015).
Yarnes, S. C. et al. Identification of QTLs for capsaicinoids, fruit quality, and plant architecture-related traits in an interspecific Capsicum RIL population. Genome 56, 61–74 (2012).
Han, K. et al. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum). DNA Res. 23, 81–91 (2016).
Elshire, R. J. et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379 (2011).
Miller, M. R., Dunham, J. P., Amores, A., Cresko, W. A. & Johnson, E. A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res. 17 (2007).
Davey, J. W. et al. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 12, 499 (2011).
Sun, X. et al. SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PloS One 8, e58700 (2013).
Hill, T. A. et al. Regions underlying population structure and the genomics of organ size determination in Capsicum annuum. Plant Genome-Us 10 (2017).
Cheng, J. et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 6, 18919 (2016).
Hulse-Kemp, A. M. et al. A HapMap leads to a Capsicum annuum SNP infinium array: a new tool for pepper breeding. Hortic. Res. 3, 16036 (2016).
Han, K. et al. QTL mapping and GWAS reveal candidate genes controlling capsaicinoid content in Capsicum. Plant Biotechnol. J (2018).
Weigel, D. & Meyerowitz, E. M. The ABCs of floral homeotic genes. Cell 78, 203–209 (1994).
Keck, E., McSteen, P., Carpenter, R. & Coen, E. Separation of genetic functions controlling organ identity in flowers. EMBO J. 22, 1058–1066 (2003).
Jofuku, K. D., den Boer, B. G., Van Montagu, M. & Okamuro, J. K. Control of Arabidopsis flower and seed development by the homeotic gene. APETALA2. Plant Cell 6, 1211–1225 (1994).
Borovsky, Y., Sharma, V. K., Verbakel, H. & Paran, I. CaAP2 transcription factor is a candidate gene for a flowering repressor and a candidate for controlling natural variation of flowering time in Capsicum annuum. Theor. Appl. Genet. 128, 1073–1082 (2015).
Liu, F. et al. PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 10, 1129–1132 (2017).
Wellmer, F. & Riechmann, J. L. Gene networks controlling the initiation of flower development. Trends Genet. 26, 519–527 (2010).
Cohen, O., Borovsky, Y., David-Schwartz, R. & Paran, I. Capsicum annuum S (CaS) promotes reproductive transition and is required for flower formation in pepper (Capsicum annuum). New Phytol. 202, 1014–1023 (2014).
Lemmon, Z. H. et al. The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation. Genome Res (2016).
Goto, K. & Meyerowitz, E. M. Function and regulation of the Arabidopsis floral homeotic gene. PISTILLATA. Genes Dev. 8, 1548–1560 (1994).
Thurow, C. et al. Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44, 100–113 (2005).
Friedrichsen, D. M. et al. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162, 1445–1456 (2002).
Qin, C. et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. P. Natl. Acad. Sci. USA 111, 5135–5140 (2014).
Wang, G. et al. Genetic mapping of anthocyanin accumulation-related genes in pepper fruits using a combination of SLAF-seq and BSA. PloS One 13, e0204690 (2018).
Liu, D. Y. et al. Construction and analysis of high-density linkage map using high-throughput sequencing data. Plos One 9, e98855 (2014).
Tao, A. et al. High-density genetic map construction and QTLs identification for plant height in white jute (Corchorus capsularis L.) using specific locus amplified fragment (SLAF) sequencing. BMC Genomics 18, 355 (2017).
Zhu, W. Y. et al. A high-density genetic linkage map for cucumber (Cucumis sativus L.): based on specific length amplified fragment (SLAF) sequencing and QTL analysis of fruit traits in cucumber. Front. Plant Sci. 7 (2016).
Hill, T. et al. Ultra-high density, transcript-based genetic maps of pepper define recombination in the genome and synteny among related species. G3-Genes Genom. Genet. 5, 2341–2355 (2015).
Paran, I. et al. An integrated genetic linkage map of pepper (Capsicum spp.). Mol. Breeding 13, 251–261 (2004).
Wen, J. & Zhang, Y.-M. Multi-QTL mapping for quantitative traits using distorted markers. Mol. Breeding 31, 395–404 (2013).
Xie, S.-Q., Wen, J. & Zhang, Y.-M. Multi-QTL mapping for quantitative traits using epistatic distorted markers. PloS One 8, e68510 (2013).
Lee, Y. R., Yoon, J. B. & Lee, J. A SNP-based genetic linkage map of Capsicum baccatum and its comparison to the Capsicum annuum reference physical map. Mol. Breeding 36, 61 (2016).
Lee, J. et al. QTL mapping for capsaicin and dihydrocapsaicin content in a population of Capsicum annuum ‘NB1’ × Capsicum chinense ‘Bhut Jolokia’. Plant Breeding 135, 376–383 (2016).
Zhang, X. et al. Candidate genes for first flower node identified in pepper using combined SLAF-seq and BSA. PloS One 13, e0194071 (2018).
Xu, S. QTL mapping can benefit from segregation distortion. Genetics (2008).
Subramanya, R. Transfer of genes for increased flower number in pepper. HortScience 18, 747–749 (1983).
Bassett, M. J. Breeding vegetable crops. (1986).
Lippman, Z. B. et al. The making of a compound inflorescence in tomato and related nightshades. PloS Biol. 6, e288 (2008).
Wang, S.-B. et al. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 6, 19444 (2016).
Kim, S. et al. New reference genome sequences of hot pepper reveal the massive evolution of plant disease-resistance genes by retroduplication. Genome Biol. 18, 210 (2017).
Doyle, J. & Doyle, J. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull 19, 11–15 (1987).
Arends, D., Prins, P., Jansen, R. C. & Broman, K. W. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26, 2990–2992 (2010).
Wen, Y. J. et al. An efficient multi-locus mixed model framework for the detection of small and linked QTLs in F2. Brief Bioinform (2018).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Acknowledgements
This work was supported by the National Key Research and Development Program (2018YFD1000800), National Natural Science Foundation of China (31572124); key project of Guangdong Science and Technology Section (2015B020202009), the key project of Guangzhou Science Technology and Innovation Commission (201508030021), and the Graduate Student Overseas Study Program from South China Agricultural University (2017LHPY018, 2018LHPY005).
Author information
Authors and Affiliations
Contributions
Z.Z.S. constructed the mapping populations, performed the genetic analysis, and wrote the paper. Z.Z.S., S.B.M, W.J.L., C.W., and H.Z.B. performed some of the field work and surveyed some traits. C.B.B., C.G.J. and C.C.M. provided valuable research ideas. L.J.J. designed and supervised the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, Z., Sun, B., Wei, J. et al. Construction of a high density genetic map of an interspecific cross of Capsicum chinense and Capsicum annuum and QTL analysis of floral traits. Sci Rep 9, 1054 (2019). https://doi.org/10.1038/s41598-018-38370-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-38370-0
This article is cited by
-
Genomic regions and candidate genes linked with Phytophthora capsici root rot resistance in chile pepper (Capsicum annuum L.)
BMC Plant Biology (2021)
-
The APETALA2 homolog CaFFN regulates flowering time in pepper
Horticulture Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.