Abstract
While only 15–25 percent of melanoma patients develop distant metastasis and die, this disease is still responsible for the majority of skin cancer-related deaths. The availability of adjuvant therapies makes the selection of high-risk patients essential. We evaluated the intratumoral expression of ten miRNAs in primary melanomas in relation to its ability to predict melanoma survival. To this end, we correlated miRNA expression in 132 cryopreserved primary and metastatic tumors with clinicopathological factors and clinical outcome. We found sequential downregulation of intratumoral expression of miR-125b, miR-182, miR-200c and miR-205 over the full spectrum of melanoma progression. Moreover, downregulation of these miRNAs occurred in primary melanomas that further disseminated to distant sites. Furthermore, miR-125b, miR-200c and miR-205 correlated as independent factors with shorter survival. Our in vitro findings demonstrate that loss of miR-205 potentiates the invasive ability of melanoma cells. We conclude that the downregulation of miR-205 in primary melanomas is an intrinsic property that might contribute to distant metastasis. In particular, the interaction of melanoma cells with the extracellular matrix is one of the key mechanisms by which miR-205 influences melanoma metastasis. In conclusion, miR-125b, miR-200c and miR-205 are useful prognostic biomarkers at the time of diagnosis to select high-risk patients.
Similar content being viewed by others
Introduction
Malignant melanoma already accounts for around 80% of all skin cancer-related deaths and its incidence is increasing worldwide1. Metastatic dissemination of cutaneous malignant melanoma is the main cause of the high mortality observed in melanoma.
There is cumulative evidence that miRNAs, which control many pathological processes acting as gene expression regulators, mediate melanoma invasion and metastasis. Here, we investigated the ability of a miRNA panel to select patients at high-risk of distant metastasis at the time of primary tumor diagnosis.
The current American Joint Committee on Cancer (AJCC) staging system for cutaneous melanoma is based on primary tumor thickness and the presence of ulceration, lymph node spread and distant metastases as determinants of prognosis2.
The identification of metastatic disease is a major factor for clinical outcome in patients with melanoma. While most melanoma patients never develop recurrent disease after excision of the primary tumor, a subset of patients (15–25%) will develop metastases and die from melanoma. However, clinical and histopathological features alone are currently unable to accurately predict clinical behavior in all melanoma cases.
Deregulation of some microRNAs and their targeted genes have been associated with the well-known hallmark biological characteristics of human cancer3,4,5,6. microRNAs (miRNAs) are endogenous small noncoding RNAs that regulate the stability or translational efficiency of target mRNAs and are involved in various biological processes, including differentiation, cell proliferation, apoptosis, transformation, invasion and migration of melanoma tumor cells7,8. miRNAs regulate the expression of more than 60% of protein coding genes and are aberrantly expressed in many human cancers contributing to the initiation and development of various types of cancer, including melanoma9. Depending on the cancer type they can function as tumor suppressor genes or oncogenes.
To date, most miRNA studies on melanoma have aimed to identify miRNAs capable of distinguishing normal tissue from melanoma tissue, or primary from metastatic disease, but few studies exist on miRNAs able to predict distant metastatic potential among primary melanomas. Our goal was to evaluate the ability to predict clinical outcome of the expression in the tumor microenvironment of human melanomas, of those miRNAs which have already been implicated in the literature in melanoma cell migration and/or invasion.
Results
miR-9, miR-125b, miR-137, miR-182, miR-200c and miR-205 expression correlate with prognostic clinicopathologic features in primary human melanoma
The clinicopathological features of the primary tumors included in our study are summarized in Table 1. We found a significant inverse correlation between miR-125b, miR-182, miR-200c and miR-205 expression and both Breslow thickness and mitotic index in primary melanomas (Spearman correlation) (Table 2). Similarly, ulcerated primary melanomas and those with vertical growth phase exhibited lower levels of miR-125b, miR-182, miR-200c and miR-205 (Mann-Whitney test) (Table 2) (Fig. 1A–D).
Correlation between miR-125b, miR-182, miR-200c and miR-205 expression and histopathologic features in primary melanomas. (A) Correlation of relative expression of miR-125b, miR-182, miR-200c and miR-205 according to Breslow thickness, (B) the number of mitoses per mm2, (C) ulceration (D) and growth phase. **p < 0.01 and *p < 0.05.
In contrast, cases with higher Breslow thickness, higher mitotic index, ulceration and vertical growth phase showed increased miR-9 and miR-137 expression (Table 2).
Differential expression profile of miR-9, miR-125b, miR-137, miR-182, miR-200c and miR-205 in the different steps of melanoma progression
Progressive loss of miR-125b, miR-182, miR-200c and miR-205 expression was found along the full spectrum of melanoma progression, from thin primary melanomas to distant metastatic tumors (miR-125b, p = 0.014; miR-182, p < 0.0001; miR-200c, p < 0.0001 and miR-205, p < 0.0001; Kruskal-Wallis test) (Fig. 2).
Comparison between the relative expression of miR-125b, miR-182, miR-200c and miR-205 along the different steps of melanoma progression. (A) Relative miR-125b, (B) miR-182, (C) miR-200c and (D) miR-205 expression in thin primary melanomas (≤1 mm), thick primary melanomas (>1 mm), in-transit metastases (IT-Met), lymph node metastases (LN-Met) and in distant metastases (D-Met).
Correlation of miR-125b, miR-182, miR-200c and miR-205 expression in primary tumors with further development of distant metastasis and melanoma specific survival
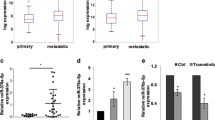
Of the primary melanomas, 28% (18/65) further metastasized to distant sites, and 89% (16/18) of these patients died of melanoma (Table 3). miR-125b, miR-182, miR-200c and miR-205 expression was significantly lower in primary tumors which further metastasized to distant sites (p < 0.001, p < 0.01, p < 0.001 and p < 0.001, respectively) a tendency which was also found in primary tumors of the patients who died from melanoma (p < 0.001, p < 0.01, p < 0.001 and p < 0.001, respectively) (Fig. 3).
Association of miR-125b, miR-182, miR-200c and miR-205 expression in primary tumors according to clinical behavior. (A) Association of miR-125b, miR-182, miR-200c and miR-205 expression in primary tumors based on the further development of distant metastasis and (B) melanoma specific death. **p < 0.01.
Correlation of miR-125b, miR-182, miR-200c and miR-205 expression in primary tumors with distant metastasis free survival and melanoma specific survival
In Kaplan-Meier survival analysis, both distant metastasis free survival (DMFS) and melanoma specific survival (MSS), were significantly longer in patients with primary melanomas showing miR-125b, miR-182, miR-200c and miR-205 expression levels above the median (Fig. 4, Table 4).
Cox multivariate analysis was performed to elucidate if these four miRNAs were independent of the main clinicopathological prognostic variables. miR-125b, miR-200c and miR-205 expression were independent of Breslow, and miR-205 independent of Breslow and ulceration (Table 5). Overall, miR-205 was the best miRNA independent predictor of melanoma specific survival.
miRNA expression in melanocytes and melanoma cell lines
Using RT-qPCR we tested the expression of the ten miRNAs in melanocytes and the following well-characterized melanoma cell lines: A375, SKMEL-147 and 451 Lu.
Our analysis showed that miR-205 (an independent prognostic factor for MSS) was the only miRNA which was not expressed in melanoma cell lines tested or melanocytes (Fig. 5). Compared with melanoma cell lines, melanocytes expressed lower miR-125b, miR-182 and miR-21 levels, did not express miR-137, and showed the highest miR-211 expression. miR-205 was therefore selected for in vitro functional assays in order to better describe the biological mechanisms affecting clinical outcome of melanoma patients.
miRNA relative expression in melanoma cell lines and in melanocytes. Quantification of the ten selected miRNAs on A375, SKMEL147 and 451 Lu human melanoma cell lines and in melanocytes by reverse transcription quantitative Real time PCR analysis (RT-qPCR). All reactions were performed in triplicate and RNU48 was used as the reference control.
miR-205 overexpression in human melanoma cells
Our goal was to determine if miR-205 overexpression in human melanoma cells is associated with changes in the proliferation, migration and invasion ability of tumor cells in vitro after transduction.
Relative mature miR-205 expression levels in A375 cells after lentiviral transduction with pmiRH-205 or pmiRH-Scr constructs were determined by TaqMan miRNA RT-qPCR. Validation of miR-205 overexpression by RT-qPCR showed high levels of mature miR-205 in pmiRH-205 transduced cells and a lack of miR-205 expression in pmiRH-Scr cells. These experiments were performed in triplicate (Fig. 6A). A very close to 100% transduction efficiency was achieved as the vast majority of cells were fluorescent (Fig. 6B).
miR-205 overexpression and inhibition in A375 human melanoma cell line. (A) Quantitative RT‐qPCR validation of miR‐205 overexpression after transfection and later infection with pmiRH-205 expression vector or pmiRH-Scr (control) vector. Data is presented as mean ± SD (n = 3). (B) Fluorescent images of GFP positive cells showing strong GFP signal on the entire cells demonstrate an efficient transduction of the miR-205 precursor or the control vector. (C) Quantitative RT-PCR validation of miR-205 inhibition of A375 overexpressing miR-205.
miR-205 inhibition of human melanoma cells overexpressing miR-205
To exclude potential undesirable effects during the overexpression procedure we specifically inhibited miR-205 in cells overexpressing miR-205. miR-205 was inhibited approximately 16-fold upon inhibition which resulted to be significant (Fig. 6C).
Functional validation of miR-205 overexpression and inhibition
We also evaluated the functional effect of miR-205 overexpression and inhibition at protein level by immunoblot densitometry after A375 cells transduction with pmiRH-205 or pmiRH-Scr and A375 overexpressing miR-205 transfected with mirVana miR-205 inhibitors or the mirVana inhibitor negative control. In melanoma, miR-205 represses expression of its direct target ZEB1. Thus, cells overexpressing miR-205 should have lower levels of ZEB1.
As expected, ZEB1 protein was significantly reduced after miR-205 overexpression when compared to the scrambled condition, thus demonstrating its efficient inhibition by miR-205 (Fig. 7A,B and Supplementary Figures 1–3).
Functional validation of miR-205 overexpression and inhibition. (A,B) ZEB1 and CDH1 relative protein levels quantification by densitometry in ImageJ (normalized to actin) of miR-205 overexpressing cells and (C,D) of miR-205 inhibited cells. (E) ZEB1 and CDH1 bands intensities as assessed by western blot (Actin: 42KDa; ZEB1: ≈70 KDa; CDH1: ≈55KDa) in miR-205 overexpression and (F) in miR-205 inhibition condition. For comparison, scrambled and miR-205 overexpression samples were loaded on the same gel and miR-205 inhibition negative control and miR-205 inhibition samples on another gel.
When miR-205 overexpression was reverted by inhibition, ZEB1 protein was again increased when compared to the negative control. Although the difference was not statistically significant, the expected tendency was clearly observed (Fig. 7C,D and Supplementary Figures 4–6). These results support the notion that miR-205 binds to ZEB1 gene and negatively regulates its expression at the protein level.
Cell proliferation assays
Although low levels of miR-205 are associated with metastatic progression, after measuring absorbance for 5 days there were no differences between proliferation of A375 cells with and without miR-205 expression (Fig. 8A).
miR-205 overexpression and miR-205 inhibition functional effects on human melanoma cells. (A) No effect on proliferation was observed between miR-205 overexpressing A375 cells versus scrambled/control cells. Data represent the mean ± SD of 4 independent experiments. (B) A representative microscopic image of crystal violet staining of the migration and invasion assays in miR-205 overexpressing A375 cells and control cells. (C and D) Transwell migration (n = 3) and matrigel invasion (n = 3) assays on A375 cells. A375 cells transduced with miR-205 had higher migratory but lower invasive potentials than the control cells. (E and F) Transwell migration (n = 3) and matrigel invasion (n = 3) assays on A375 cells transfected with miR-205 inhibition negative control and miR-205-5p inhibitor. miR-205 inhibited cells migrated less but had higher invasive ability. Data represent the mean ± SD of 3 independent experiments. **p < 0.01, *p < 0.05.
Migration and Invasion Assays
miR-205 was overexpressed and then inhibited in A375 cells and migration and invasion assays performed for both conditions. After 48 hours, there were more miR-205 overexpressing cells that had migrated to the Transwell insert lower chamber in comparison with cells without miR-205 overexpression (p < 0.01). However, in the invasion assay, cells that expressed miR-205 invaded less than cells with no expression of the miR-205 (p < 0.05) (Fig. 8B–D).
As expected, after inhibition of miR-205 overexpression, opposite results were observed. Inhibited cells migrated less than miR-205 overexpressing cells (p = 0.011) whereas in the invasion assay cells with inhibited miR-205 overexpression invaded more although values did not reach significance (p = 0.325) (Fig. 8E,F).
Discussion
Defining an accurate prognosis for patients with primary melanoma is an essential clinical goal. Presently, patient selection is based exclusively on the histopathological features. However, it is well known that patients with clinicopathologically analogous primary tumors can have different clinical outcomes. In addition to histopathologic characteristics, new molecular biomarkers are needed to better understand, diagnose, and treat melanoma patients. Moreover, only patients with advanced disease (distant metastasis and or unresectable regional metastases) currently benefit from immunotherapy and inhibitory molecules treatment, although their administration in high-risk patients before progression is under consideration.
It is widely accepted that miRNAs are dynamically involved in early and late events driving melanoma progression. Numerous studies highlight their potential as diagnostic and prognostic markers, as well as therapeutic targets in melanoma10,11,12,13,14,15,16,17,18. In this regard, van Kempen et al.19 found 11 miRNAs whose expression was associated with tumor thickness, and that loss of miR-200a, miR-200c and miR-203 expression was present at the invasive front in primary tumors. In particular, four miRNAs (miR-200b, miR-200c, miR-203, and miR-205) correlated with thickness; two of which (miR-200c and miR-205) maintained the same direction as in our data.
We hypothesize that alterations in the expression of certain miRNAs in the tumor microenvironment of primary melanomas may predict distant metastasis. We evaluated the prognostic value of the expression of ten miRNAs, which have already been implicated in melanoma cell migration and/or invasion, in primary cutaneous melanoma tissue, particularly for its ability to predict distant metastasis and, consequently, melanoma specific survival. For this purpose, we correlated miRNA expression with the clinicopathological prognostic factors and, more importantly, with the clinical outcome of the patients.
miR-21, miR-125b, miR-150, miR-155, miR-205 and miR-211 are dysregulated in melanoma10. We found that miR-125b, miR-182, miR-200c and miR-205 expression was higher in primary tumors than in melanoma metastases. In fact, it has been suggested that miR-200c and miR-205 downregulation in tumor cells is an essential early step in the development of metastasis because of its important role as promotor of cancer progression20 at least in part through the regulation of E-cadherin expression during the epithelial-mesenchymal transition targeting its transcriptional repressors ZEB1 and ZEB220. miR-205 has been proposed as a tumor suppressor microRNA in melanoma21. Dar et al. reported that miR-205 mediates its effects on melanoma cells partially via suppression of the E2F family of transcription factors E2F1 which play a role in the control of cell cycle and the AKT pathway in melanoma cell lines22.
In our series, downregulation of miR-125b, miR-182, miR-200c and miR-205 occurs sequentially over the full spectrum of melanoma progression. Moreover, we found a lower intratumoral expression of these four miRNAs in primary melanomas that further disseminated to distant sites compared with those that did not metastasize. Similarly, by comparing primary tumors that metastasized to regional lymph nodes with those that did not, Glud and coworkers23 identified nine miRNAs as differentially expressed, and miR-125b was downregulated in those with positive lymph nodes. According to these authors, the overexpression of miR-125b, a regulator of melanogenesis and differentiation of melanocytes24, was associated with senescence in melanoma cells, and decreasing levels of apoptosis when miR-125b was inhibited in the primary cell lines25,26. Based on these studies, Kappelmann et al.27 reported that miR-125b controlled melanoma progression by directly targeting c-Jun, as well as the downregulation of miR-125b expression in melanoma cell lines. MLK3 (mixed lineage kinase 3) proliferation and invasion promoting proteins, are also miR-125b targets in melanoma28.
A reduction in proliferation and migration has been reported when the expression of miR-125b was restored in the cell lines, suggesting that miR-125b expression had a tumor suppressor function in melanoma27. Moreover, a recent study showed a decreased expression of miR-125b in primary tumors with positive sentinel lymph nodes, and concluded that miR-125b exerted a negative metastatic regulation on melanoma cell invasion in vitro and in vivo by targeting ITGA9, and therefore inhibiting the epithelial-mesenchymal transition in melanoma29.
Our in vitro results in melanoma cells with and without miR-205 functional expression demonstrate that the loss of miR-205 potentiates the matrigel invasive ability of melanoma cells, although interestingly, neither proliferation nor migration was increased. Since invasion assays best reproduce the in vivo environment, in which cancer cells must interact constantly with the extracellular matrix, our findings reveal that tumor cell interaction with the extracellular matrix is critical to miR-205 inhibition of metastatic dissemination. In this regard Liu et al. found that miR-205 downregulation did not modify proliferation of melanoma cells but increased both invasion and migration30. In contrast, other authors have found that miR-205 either suppresses melanoma cell proliferation22 or makes no difference related to invasion31.
The transcriptomic subtypes of TCGA classification of cutaneous melanoma showed a distinctive miRNA expression profile32. However, among our selected miRNAs, only miR-125b was found to be downregulated in the worse prognostic “keratin expressor” subtype of this classification. On the other hand, upper levels of miR-125b were found in the “MITF-low” subgroup associated with better survival. These results support our findings, since miR-125b significantly correlated with worse DMFS and MSS in our cohort. miR-182, miR-200c and miR-205 expression levels did not correlate with any TCGA transcriptomic subtypes. In an in silico analysis using the TGCA database, Lohcharienkal et al.33 also found a decreased expression of miR-200c and miR-182 in primary vs metastatic melanomas. These findings agree with our observations, since miR-200c and miR-182 expression were significantly decreased in the different stages of melanoma progression. However, the above study did not evaluate the clinical implications of these differences.
In conclusion, downregulation of tumor suppressive miR-125b, miR-200c and miR-205 in primary melanomas is an intrinsic property of primary tumors that might contribute to distant metastasis, and therefore to reduced survival. One of the mechanisms by which miR-205 influences melanoma metastasis and survival seems to be related to the interaction of melanoma cells with the extracellular matrix.
Overall, these findings reveal a key role for these three miRNAs in melanoma metastasis. Moreover, their intratumoral expression in primary tumors may serve as effective prognostic molecular biomarkers in order to select high-risk melanoma patients. Furthermore, potentiating the expression levels of these three miRNAs, and particularly that of miR-205, might have a potential therapeutic value in melanoma patients at the time of diagnosis of the primary tumor, by preventing metastatic dissemination and improving survival in the context of personalized medicine.
Methods
Human melanoma tissues
132 cryopreserved tumor specimens from patients with cutaneous malignant melanoma were selected for this study. Samples comprised primary melanomas (n = 65) and melanoma metastases (n = 67), of which 28 were in-transit, 24 regional lymph node and 15 distant metastases (4 skin, 3 lung, 2 brain, 2 soft tissue, 2 subcutaneous tissue, 1 bone, 1 liver). All tumor specimens were collected at the time of surgery at the Department of Anatomic Pathology, Hospital Clínico Universitario, Valencia, Spain, from November 2002 to December 2014.
Primary tumor parameters relevant to this study included Breslow thickness, mitotic index, ulceration, growth phase, location, gender, stage and histological type. Clinical follow-up, with particular emphasis on the development of distant metastases and melanoma mortality, ranged from 6.5 to 156 months (mean 71, median 68 months). All tumors were classified according to the 2017 American Joint Committee on Cancer (AJCC) staging system.
Both primary and metastatic tumor specimens were manually macrodissected to ensure maximum tumor tissue content. For primary melanomas, a tumor slice immediately adjacent to the thickest area of the tumor was selected for RNA extraction, and was immediately frozen in liquid nitrogen and stored at -80 °C. The remaining fresh tumor tissue from each case was formalin-fixed and paraffin-embedded for routine diagnosis. This protocol was approved by the Ethical and Scientific Committees, Hospital Clínico Universitario, Valencia, and all their guidelines were followed. Medical records from all patients were reviewed and clinical follow-up locked in December 2016. All patients provided written informed consent.
Human melanoma and melanocyte cell culture
The following well-characterized melanoma cell lines have been used: A375, SKMEL-147 and 451 Lu. The latter was kindly provided by Dr. Eva Hernando (New York University Langone Medical Center, NY), SK-MEL-147 by Dr. Marisol Soengas (CNIO Melanoma Group) and human epidermal melanocytes by Dr. Julián Carretero (Universitat de València).
Human melanocytes were cultured in 254CF medium supplemented with calcium chloride at a final concentration of 0.2 mM, with Human Melanocyte Growth Supplement (Cascade Biologics Inc., Gibco), penicillin and streptomycin. Medium was changed daily.
Melanoma cells were grown in Dulbecco’s Modification of Eagle’s Medium (DMEM) with glucose and L-glutamine supplemented with 10% FBS, penicillin and streptomycin (Gibco) and incubated in a humidified incubator at 37 °C and 5% CO2. To avoid cultures above 80% confluency, subcultures were made regularly with 0.25% trypsin–EDTA (Gibco). All cells were routinely tested for Mycoplasma contamination using MycoAlert™ mycoplasma detection kit (Lonza).
RNA extraction
mirVana miRNA Isolation Kit (Ambion) was used to perform total RNA extraction from patient melanoma tissue and cultured cells following the manufacturer’s recommendations. The procedure for tissues differed from that of cultured cells only in the cell lysis step.
Melanoma tissues were immediately disaggregated with a pre-chilled scalpel into tiny portions and were mechanically homogenized in 600 µl of Lysis/Binding buffer using the TissueLyser LT system (Qiagen). For cultured cells, 600 µl of Lysis/Binding solution were added and samples vortexed. All the following procedures were carried out in the same manner and according to the manufacturer’s protocol with minor modifications. RNA was quantified using Nanodrop ND-1000 Spectrophotometer (Thermo Scientific) and stored at -80 °C.
miRNA quantification
From human samples and from cultured cells, relative quantification of mature microRNAs was carried out by reverse transcription quantitative Real time PCR (RT-qPCR). Reverse transcription (RT) was performed using TaqMan MicroRNA Reverse Transcription Kit using RNase inhibitor (Lifetechnologies). In 45-µl reactions, 200 ng of total RNA was converted to cDNA. RT reactions were multiplexed by customizing RT primer pool with miRNA-specific RT primers of interest following manufacturer’s recommendations34,35. The RT primers pooled were the provided along with the TaqMan MicroRNA Assays (Applied Biosystems) that are listed in Table 6.
In brief, 1 µl of cDNA was used in a 10-μl qPCR reaction by adding TaqMan Universal Master Mix II, no UNG and TaqMan MicroRNA Assays for target miRNAs (Lifetechnologies). Small nuclear RNU48 was used as endogenous control. All reactions were performed in triplicate in 384-well plates on a 7900 HT Fast Real- Time PCR system (Lifetechnologies). Both, RT and qPCR negative controls were included for each assay.
Samples that showed Ct median values for the endogenous reference out of the range (from 17 to 23) were classified as not appropriate for normalization and, consequently, excluded from further qPCR analysis. For the relative quantification of miRNA expression, the Ct method was used and the results were analyzed using Expression Suite software (Lifetechnologies).
miRNA overexpression
Two different miRNA expression constructs: a lentiviral construct and a non-viral expression plasmid were tested for efficiency of miR-205 overexpression and the most efficient (the lentiviral construct) was selected for further experimentation.
Lentiviral constructs for miR-205 (pmiRH-205) and scrambled negative control (pmiRH-Scr) were purchased from System Biosciences. The construct also carries a fluorescent marker (GFP) to monitor positive cells for transfection and transduction. First, lentiviral particles were produced by transient transfection of HEK293T cells with lentiviral expression constructs using Lipofectamine 2000 in Opti-MEM (Invitrogen).Lentiviral supernatant was filtered using 0.45 µm filters at 36 hours post-transfection and stored at −80 °C.
A375 target cells were seeded at a density that produced approximately 70% confluence in 24 hours and incubated overnight preceding infection. Medium was replaced with filtered 1:4 diluted viral supernatant containing 4 µg/ml polybrene (Sigma), and cells were then incubated for 4 hours before removing the viral supernatant followed by replacement with fresh medium. Transduction efficiency was checked for GFP expression. miR-205 overexpression was validated by RT-qPCR as detailed in miRNA quantification section.
miRNA transient inhibitor transfection
Downregulation of miR-205 activity was achieved using mirVana inhibitors. Optimal transfection conditions that resulted in maximum miRNA inhibitor-mediated activity with minimal cytotoxicity were determined and maintained across experiments and controls. mirVana miRNA Inhibitors (negative control 4464076 and hsa-miR-205-5p ID MH11015) were transfected in 6-well plates for a final concentration of 50 nM using Lipofectamine (Lifetechnologies). Maximal inhibitor activity was achieved at 24 hours post-transfection.
Protein isolation
Cell lysates were prepared in RIPA buffer freshly supplemented with sodium orthovanadate and protease inhibitors cocktail (Sigma) on ice. After centrifugation for 30 minutes at 14000 rpm at 4 °C, the supernatant was collected and protein concentration was determined by Lowry method reading absorbance at 660 nm on a multilabel plate reader (VICTOR™ X3 Multilabel Plate Reader, PerkinElmer). Standard curves were generated with BSA. Proteins from inhibition experiments were isolated 24 hours post-transfection.
Western blotting
Cell lysates 50 µg of protein per lane) were resolved in Tris/glycine SDS-PAGE gradient (4–20%) gels (Mini-PROTEAN TGX Stain-Free gels, Bio-Rad) and transferred to PVDF membranes by semi-dry blotting (Bio-Rad Trans-Blot Turbo system). Membranes were blocked for 1 h at room temperature with 5% milk in TTBS. Membranes were probed with primary antibodies against AREB 6/ZEB1 (Abcam ab181451), E-Cadherin (CDH1) (Abcam ab40772) and as loading control b-Actin-Peroxidase (Sigma A3854) overnight at 4 °C, followed by appropiate HRP-conjugated secondary antibodies (Cell Signaling Anti-mouse IgG HRP-linked Antibody and Peroxidase AffiniPure Goat Anti-Rabbit IgG) except for actin.
Immunolabeled proteins were detected by incubation with the ECL-based SuperSignal West Pico PLUS Chemiluminescent detection Substrate (Thermo Scientific) using the ImageQuant LAS 4000 (GE Healthcare Life Sciences) imaging system. Sample and control band densities were quantified and compared with ImageJ. Data was normalized to actin.
Four replicates of scrambled and miR-205 overexpression were loaded on the same gel. For miR-205 inhibition negative control and miR-205 inhibition, three replicates per condition were loaded on the same gel. In all cases, all the replicates were used for each quantification in ImageJ. Three representative bands per condition were cropped from different parts of the same gel for the three genes per experiment. Full-length blots are included in Supplementary Figures 1–6.
Proliferation assays
A375 cells were seeded at low density (1 × 103 cells/well in 100 µl) in a 96-well plate. Before seeding, cell viability was analyzed by the trypan blue exclusion method. Cells were then fixed, washed and stained with Crystal Violet. Cells were destained with 15% acetic acid and absorbance was measured at 590 nm on a VICTOR™ X3 Multilabel Plate Reader (PerkinElmer). The absorbance value measured after 24 hours was used as a control and the relative absorbance value was calculated every 24 hours for 5 days. The experiment was repeated four times.
Migration and invasion Assays
A 300 µl suspension of 4 × 104 A375 transduced melanoma cells was added to Transwell inserts (Corning) uncoated or coated with Matrigel (Corning) diluted 1:40 in coating buffer for migration and invasion assays, respectively. Matrigel coated inserts were incubated for 2 hours at 37 °C. After coating, cells were seeded in the inner chamber in serum-free DMEM medium, and 500 µl of DMEM medium containing 10% FBS was added to the outer chambers. At counting for seeding, cell viability was analyzed by the trypan blue exclusion method. An equivalent volume of cell suspension was transferred to empty wells as cell input controls.
After forty-eight hours, cells were fixed (1% Glutaraldehyde in PBS), washed in PBS and stained with 0.5% Crystal Violet followed by extensive washing with H2O. Non-migrated or invasive tumor cells remaining on the top-side of the porous membrane were removed with a cotton swab. Cells that had migrated or invaded to the downside were photographed and scored by imaging and counting four fields per insert (inserts in triplicate per condition). All the experiments were repeated in triplicate. The migration and invasion assays were performed for transient transfection experiments with inhibitors as described above 24 hours post-transfection.
Statistical analysis
Data plotting and statistical analysis were conducted using SPSS software package V.17.0 (SPSS Software, Inc.) and GraphPad Prism V.6.01 (GraphPad Software, Inc.).
Two-tailed Mann-Whitney test was used to analyze the association between miRNA expression and categorical clinicopathological parameters (two categories) while Kruskal-Wallis rank-sum test was used to compare categorical parameters defined by more than two different groups. The correlation between miRNA expression and continuous clinicopathological variables (Breslow thickness and mitotic index) was assessed by Spearman correlation.
Survival analysis was performed using Kaplan-Meier curves (Log-rank test). A Cox proportional hazard model was constructed using stepwise selection to identify independent predictors of clinical outcome, considering Hazard Ratios (HR), 95% CI and p values.
Distant metastasis free survival (DMFS) and melanoma specific survival (MSS) were defined as the period from the date of surgical excision of the primary melanoma to the date distant metastasis occurred or last follow-up (censored), or death from melanoma (event), respectively. For all statistical tests, a p value of less than 0.05 was considered statistically significant.
Data Availability Statement
The authors will make materials, data and associated protocols promptly available to readers upon publication in Scientific Reports without undue qualifications in materials transfer agreements.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2017. CA Cancer J Clin 67, 7–30 (2017).
Gershenwald, J. E. et al. Melanoma of the skin in AJCC Cancer Staging Manual, 8th ed (ed. Amin, M. B.) 563–585 (Springer, 2017).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Dalmay, T. & Edwards, D. R. MicroRNAs and the hallmarks of cancer. Oncogene 25, 6170–6175 (2006).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Bennett, P. E. et al. miR in melanoma development: miRNAs and acquired hallmarks of cancer in melanoma. Physiol Genomics 45, 1049–1059 (2013).
Bartel, D. P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 116, 281–297 (2004).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Martinez-Rodriguez, M., Thompson, A. K. & Monteagudo, C. High CCL27 immunoreactivity in “supratumoral” epidermis correlates with better prognosis in patients with cutaneous malignant melanoma. J Clin Pathol 70, 15–19 (2017).
Latchana, N., Ganju, A., Howard, J. H. & Carson, W. E. MicroRNA dysregulation in melanoma. Surg Oncol 25, 184–189 (2016).
Mirzaei, H. et al. MicroRNAs as potential diagnostic and prognostic biomarkers in melanoma. Eur J Cancer 53, 25–32 (2016).
Jayawardana, K. et al. Identification, Review, and Systematic Cross-Validation of microRNA Prognostic Signatures in Metastatic Melanoma. J Invest Dermatol 136, 245–254 (2016).
Sun, V., Zhou, W. B., Majid, S., Kashani-Sabet, M. & Dar, A. A. MicroRNA-mediated regulation of melanoma. Br J Dermatol 171, 234–241 (2014).
Völler, D., Ott, C. & Bosserhoff, A. MicroRNAs in malignant melanoma. Clin Biochem 46, 909–917 (2013).
Glud, M. & Gniadecki, R. MicroRNAs in the pathogenesis of malignant melanoma. J Eur Acad Dermatol Venereol 27, 142–150 (2013).
Kunz, M. MicroRNAs in melanoma biology. Adv Exp Med Biol 774, 103–120 (2013).
Segura, M. F., Greenwald, H. S., Hanniford, D., Osman, I. & Hernando, E. MicroRNA and cutaneous melanoma: from discovery to prognosis and therapy. Carcinogenesis 33, 1823–1832 (2012).
Bonazzi, V. F., Stark, M. S. & Hayward, N. K. MicroRNA regulation of melanoma progression. Melanoma Res 22, 101–113 (2012).
van Kempen, L. C. et al. Loss of microRNA-200a and c, and microRNA-203 expression at the invasive front of primary cutaneous melanoma is associated with increased thickness and disease progression. Virchows Arch 461, 441–448 (2012).
Gregory, P. A. et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10, 593–601 (2008).
Hanna, J. A., Hahn, L., Agarwal, S. & Rimm, D. L. In situ measurement of miR-205 in malignant melanoma tissue supports its role as a tumor suppressor microRNA. Lab Invest 92, 1390–1397 (2012).
Dar, A. A. et al. miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem 286, 16606–16614 (2011).
Glud, M. et al. Downregulation of miR-125b in metastatic cutaneous malignant melanoma. Melanoma Res 20, 479–484 (2010).
Kim, K. H. et al. Novel inhibitory function of miR-125b in melanogenesis. Pigment Cell Melanoma Res 27, 140–144 (2014).
Glud, M. et al. MicroRNA miR-125b induces senescence in human melanoma cells. Melanoma Res 21, 253–256 (2011).
Nyholm, A. M. et al. miR-125b induces celular senescence in malignant melanoma. BMC Dermatol 14, 8 (2014).
Kappelmann, M., Kuphal, S., Meister, G., Vardimon, L. & Bosserhoff, A. K. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 32, 2984–2991 (2013).
Zhang, J. et al. MLK3 promotes melanoma proliferation and invasion and is a target of microRNA-125b. Clin Exp Dermatol 39, 376–384 (2014).
Zhang, J. et al. MicroRNA-125b suppresses the epitelial-mesenchymal transition and cell invasion by targeting ITGA9 in melanoma. Tumour Biol 37, 5941–5949 (2016).
Liu, S. et al. Loss of microRNA-205 expression is associated with melanoma progression. Lab Invest 92, 1084–1096 (2012).
Xu, Y., Brenn, T., Brown, E. R., Doherty, V. & Melton, D. W. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour supressors. Br J Cancer 106, 553–561 (2012).
Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell. 161, 1681–1696 (2015).
Lohcharoenkal, W. et al. Genome-Wide Screen for MicroRNAs Reveals a Role for miR-203 in Melanoma Metastasis. J Invest Dermatol 138, 882–892 (2018).
Chen, C. et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33, 1–9 (2005).
Le Carré, J., Lamon, S. & Léger, B. Validation of a multiplex reverse transcription and pre-amplification method using TaqMan MicroRNA assays. Front Genet 5, 413 (2014).
Acknowledgements
Supported by grants, PROMETEO II/2015/009 (to CM) from Generalitat Valenciana, Spain; PI13/02786 and PI17/02019 (to CM) from Instituto de Salud Carlos III, Spain; FEDER European funds, and Proyecto Everest from the Asociación Española Contra el Cancer (AECC). We want to particularly acknowledge the patients and the INCLIVA BioBank (PT13/0010/0004) integrated in the Spanish National Biobanks Network for its collaboration.
Author information
Authors and Affiliations
Contributions
B.S.-S. and C.M. conceived of the study, designed and interpreted experiments. B.S.-S. performed experiments and analyzed data with assistance from J.F.G.-M., C.M.-C. and A.M. C.M. and L.T. collected patient tissue samples, evaluated the clinicopathological parameters and performed the clinical follow-up. B.S.-S. and C.M. wrote the manuscript. All authors commented on and accepted the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sánchez-Sendra, B., Martinez-Ciarpaglini, C., González-Muñoz, J.F. et al. Downregulation of intratumoral expression of miR-205, miR-200c and miR-125b in primary human cutaneous melanomas predicts shorter survival. Sci Rep 8, 17076 (2018). https://doi.org/10.1038/s41598-018-35317-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-35317-3
Keywords
This article is cited by
-
RETRACTED ARTICLE: MicroRNA-205-5p inhibits skin cancer cell proliferation and increase drug sensitivity by targeting TNFAIP8
Scientific Reports (2021)
-
Reconstruction of lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveals functional lncRNAs in skin cutaneous melanoma
BMC Cancer (2020)
-
Transcriptomic identification of miR-205 target genes potentially involved in metastasis and survival of cutaneous malignant melanoma
Scientific Reports (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.