Abstract
While many tropical plants have been adapted to temperate cultivation, few temperate plants have been adapted to the tropics. Originating in Western Europe, Brassica oleracea vernalization requires a period of low temperature and BoFLC2 regulates the transition to floral development. In B. oleracea germplasm selected in Taiwan, a non-vernalization pathway involving BoFLC3 rather than BoFLC2 regulates curd induction. In 112 subtropical breeding lines, specific haplotype combinations of BoFLC3 and PAN (involved in floral organ identity and a positional candidate for additional curd induction variation) adapt B. oleracea to high ambient temperature and short daylength. Duplicated genes permitted evolution of alternative pathways for control of flowering in temperate and tropical environments, a principle that might be utilized via natural or engineered approaches in other plants. New insight into regulation of Brassica flowering exemplifies translational agriculture, tapping knowledge of botanical models to improve food security under projected climate change scenarios.
Similar content being viewed by others
Introduction
Brassica oleracea (CC genome, 2n = 18), originating from coastal areas of Western Europe, has been domesticated and selected for diverse morphological variation to form important crops including broccoli, Brussels sprouts, cabbage, cauliflower, kale, and others. Broccoli (var. italica), cultivated for its thickened edible inflorescence1, has also gained attention for containing rich anticancer compounds, glucosinolates2 and demand is increasing globally. However, broccoli production is restricted to limited areas or cool seasons because its flowering is triggered by facultative vernalization and curds properly form under low temperature. High temperature impedes differentiation of floral meristems and results in uneven-sized floral buds3,4. To adjust flowering time to optimize broccoli production will support food security and food resilience. Characteristics of non-vernalization and heat tolerance are compulsory for broccoli cultivation under global warming, and for extending cultivation to subtropical zones.
Flowering time (FTi), the consequence of the transition from the vegetative stage to the reproductive stage of growth, is genetically regulated; is highly responsive to environmental cues; and is a prime trait for plant adaptation to diverse geographic regions and seasons5,6. In addition, FTi is of central importance to determining crop cultivation and harvest seasons to optimize yield and quality. More than 300 flowering genes have been identified in the model plant, Arabidopsis thaliana, and genetically characterized into five main pathways, converging on two major floral integrators, FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), which transition meristem identity from shoot apical meristems to inflorescence meristems5,7,8,9. The gene expression of both FT and SOC1 is supressed by FLOWERING LOCUS C (FLC), which expression is reduced by vernalization, a long duration of low temperature10. Since Arabidopsis and Brassica are members of the Brassicaceae family, their genetic pathways of flowering tend to be conserved, with orthologous genes for flowering time/curd initiation time, and substantial vernalization needed to promote flowering11,12,13. However, flowering genes, like other Brassica genes, occur in multiple copies as a result of whole genome triplication after its divergence from a common ancestor shared with Arabidopsis14. For instance, only one FLC gene was identified in Arabidopsis versus four, four, and five in B. oleracea, B. rapa (AA genome, 2n = 20), and B. napus (AACC, 2n = 38), respectively12,15,16. Presence/absence variation of flowering genes increases genetic diversity for adaptation to a wide range of climatic zones and latitudes17,18.
To identify genes/QTLs affecting flowering time is important to investigating mechanisms underlying the vegetative-reproductive transition in plants, and also can provide information and tools to accelerate marker-assisted selection (MAS) to breed new varieties and broaden cultivation seasons and areas. QTL mapping combined with a candidate gene approach to isolate homologous flowering genes is popular in Brassica because of colinearity with Arabidopsis13,19,20. Numerous studies using different mapping populations, revealed conserved QTLs across different populations and environments12,13,21,22,23,24,25,26. The most conserved QTL on chromosome O2 was inferred to be BoFLC2, a vernalization responsive gene in dosage dependent regulation of flowering time with different BoFLC2 alleles accounting for flowering time variation11,12,23,27.
Early curd induction and heat tolerance are important traits for broccoli and cauliflower breeding in Taiwan and other Southeast Asian countries28, and a breeding program to develop heat-tolerant broccoli hybrids adapted to the eastern United States was launched in the early 1990s29. QTLs conferring days to curd initiation (DCI) are influenced by ambient temperature, as exemplified by only 2 QTLs were consistently detected in a doubled haploid (DH) population and a commercial diversity panel of cauliflower, respectively, at several temperature regimes22,26. BoVRN2 and BoFLC2 were not related to temperature-regulated curd induction, and suppression of BoCAL, BoAP1-a, and BoLFY or failure to suppress BoTFL1 could maintain arrest of the inflorescence meristem but not floral initiation26,30. The genetic flowering pathway of curd induction and formation in response to temperature, especially to high temperature, is still obscure. We used advanced breeding lines selected under the subtropical environment, relative hot and high humidity, to unveil more genes/QTLs provide new insights into flowering time and curd formation which can be applied to the breeding of broccoli and other Brassica crops.
Results
QTL conferring days to curd induction and curd quality
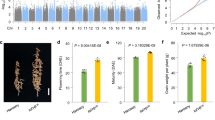
From 112 breeding lines selected under high temperature and humidity by Known-You Seed Co. in Taiwan, an early-maturing kale-derived broccoli (BLM29) was chosen for crossing with a late elite broccoli (BLM25), with the parents and 188 F2:3 families evaluated at Tainan, Taiwan (23.079337°N, 120.295377°E) under daily temperatures ranging from 24.5–30.4 °C with a 28.8 °C average. Days to curd induction (DCI) of the parents were 72 and 103 days, while F2 progeny averaged 80.2 (±5.6) days and ranged from 59.1 to 117.0 days, with three individuals exhibiting earlier DCI than BLM29 (Fig. 1a). Curd quality (CQ) was rated from 1 to 4 based on the abundance of leafy bracts between inflorescence internodes (Fig. 1b). While late-flowering BLM25 was heat sensitive and failed to initiate curds, early-flowering BLM29 formed curds with leafy bracts, rated CQ degree 3. The average CQ of the 177 survivors was 3.1 (±0.37, Fig. 1a), with only one having ideal CQ of 1, but 60 with better CQ than BLM29.
The Phenotypic evaluation and QTL intervals of days to curd induction (DCI) and curd quality (CQ). (a) The frequency distributions of DCI and CQ in the F2 population. The DCI and CQ of BLM 29 and BLM 25 are indicated by solid (▼) and open (▽) triangles, respectively, and the means of DCI and CQ of the F2 population are indicated by arrows (⇧). BLM25 with long DCI failed to initiate curd formation. (b) The evaluation of curd quality. The curd quality of broccoli was scored as 1 to 4 according to floral development and abundance of bracts in the leafy curd, from left to right. (c) The QTL intervals are indicated by bars and whiskers for 90 and 99% likelihood, respectively, and solid and striped bars are for QTLs conferring DCI and CQ, respectively. SSR markers are indicated with prefixes of BnGMS, Ol, BoGMS, fito, BRMS, Na, CB, BRAS, SORA, BrGMS, and CHT, and IP markers are indicated with prefix of At.
Using a genetic map of 60 well-spaced DNA markers forming nine linkage groups spanning 622.2 cM (Fig. 1c; the primary genetic linkage map with 126 markers is Supplementary Fig. 1), DCI QTLs were identified on chromosomes O3, O4, O6, and O7, explaining 4.03% to 28.54% of phenotypic variance (PV) (Table 1; Fig. 1c). Significant digenic interactions were detected for qDCI-6 with both qDCI-3 and qDCI-7. A full model, including 4 QTLs and two digenic interactions, explained 55.28% of PV (Table 1). The alleles of early parent BLM29 promoted DCI by 3.81 and 2.07 days for qDCI-6 and qDCI-7, respectively; postponing DCI by 3.80 and 1.78 days for qDCI-3 and qDCI-4, respectively. CQ QTLs were detected on chromosomes O2, O3, and O6 (Table 1; Fig. 1c). The major QTL, qCQ-6, mapped to the fito203/CHT104 interval, showing an additive −0.44 degree reduction of curd leafiness by the BLM29 allele, and a full model with the 3 identified QTLs explained 33.02% of PV. The BLM29 allele for qCQ-2 and qCQ-6, but not qCQ-3, increased curd quality.
The two major DCI and CQ QTLs coincided with one another, mapping to chromosomes O3 and O6 (Table 1; Fig. 1c), implying that these two QTL pairs might each be due to single genes with pleiotropic effects, or different tightly linked genes. The additive effects of these two QTL pairs contrasted in direction–elite BLM25 conferred advantageous alleles qDCI-3 and qCQ-3 for early curd induction with good curd formation. BLM29, with early DCI and heat-insensitivity, contributed allele(s) of qDCI-6 promoting curd induction by 3.81 days; and qCQ-6 reducing curd leafiness by 0.44 degrees.
To validate their effects, qDCI-6/qCQ-6 alleles of BLM29 were introgressed into BLM25–four BC3F1 individuals possessed 91.1%, 94.5%, 94.5%, and 94.5% of BLM25 DNA marker alleles and horticultural traits closely resembling BLM25 but with DCIs of 42, 42, 50, and 50 days, much earlier than BLM25 (64 days) (Supplementary Figs 2, 3). In 113 BC2F2 families, the average DCI of BLM25 homozygotes, 48.9 (±4.6), was significantly later than BLM29 homozygotes and heterozygotes, 39.8 (±5.2) and 42.3 (±5.2) (Supplementary Table 1). In 87 BC3F2 families, the DCIs of BLM25 homozygotes and BLM29, 63.1 (±6.8) and 59.1 (±5.6), also differed significantly. BLM29 homozygotes of qDCI-6 had earlier DCI than BLM25 homozygotes.
Identification of candidate genes
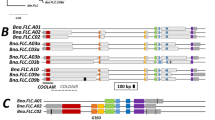
The two regions containing QTL pairs each contained eight genes with predicted functions related to flowering time and floral organ differentiation (Supplementary Table 2), a subset of which showed striking functional differences between the parents after sequence alignment analyses of resequencing data of the candidate genes. In the At5g7910-CHT20 region containing qDCI-3 and qCQ-3, only Bol008758, annotated as FLOWERING LOCUS C (FLC), showed allelic variation between BLM29 and BLM25 (Fig. 2). BLM29 had the same gene sequence as the B. oleracea var. capitata reference genome but had 244- and 678-bp indels upstream14; while 3 SNPs caused non-synonymous substitutions in BLM25 (Fig. 2). Bol008758 corresponds to BoFLC3 according to the linkage maps and functions of FLC paralogs in B. oleracea13,31. The BoFLC3 sequences of BLM29 and BLM25 were identical to those of B. oleracea var. alboglabra (Chinese kale, AM231518) and var. capitata (cabbage, AY306125).
Schematic structures of three candidate genes for DCI and CQ. BoFLC3 is the most likely candidate gene for qDCI-3/qCQ-3, and PAN and VRN2 are the candidate genes for qDCI-6/qCQ-6. The sequences of loci Bol008758, Bol02400, and Bol032823 corresponding to BoFLC3, PAN, and VRN2, respectively were retrieved from the BolBase database. AM231518 and AY306125 are two different alleles of BoFLC3 from Chinese kale and cabbage, respectively. Gene structure with the important domains of these three genes are indicated. SNPs causing non-synonymous amino acids are indicated. G1, G2, and G3 are the haplotypes identified from 112 broccoli breeding lines according to the sequence variation. The exons and introns are represented by solid boxes and lines, respectively. The solid triangle (▲), double slash (//), and solid circle (●) indicate insertion, deletion, and pre-mature stop codon, respectively. BoFLC3 is comprised of 7 exons and 6 introns; and variations of insertion, deletion, and substitution were found in introns 1 and 6; and exons 2, 4, and 6. PAN is comprised of 10 exons and 9 introns for which nonsynonymous substitutions were detected on exons 1 and 8 and one early stop codon on exon 10. VRN2 is comprised of 14 exons and 13 introns, two nonsynonymous substitutions were found on exons 6 and exon 8 and one nonsynonymous substitution and two indel amino acids were detected on exon14. The cis-elements on the deletion fragments of BoFLC3 upstream were predicted by PlantPAN 2.0 using Arabidopsis database46.
In the fito23-fito36 interval containing qDCI-6 and qCQ-6, the BLM29 allele of PAN (PERIANTHIA, Bol024000), a bZIP-transcription factor required for AGAMOUS activation in Arabidopsis flowers32, has the same amino acid sequence as Bol024000; BLM25 has three non-synonymous substitutions (Fig. 2). The BLM25 allele of VRN2 (VERNALIZATION2, Bol032823), a vernalization-responsive repressor of FLC, was identical to Bol032823, cabbage VRN2; BLM29 had two non-synonymous substitutions and two additional amino acids on exon 14. The other six candidate genes lacked non-synonymous substitution.
BLM29 and BLM25 showed significantly different expression levels of BoFLC3 in the vegetative leaf at the 4-leaf stage (VL); the latest leaf at curd forming (CL); and the peduncle bract at harvest stage (PB); PAN in floral bud at harvest stage (FB: Fig. 3); and VRN2 in PB. BoFLC3 and PAN expression levels were relatively low–VRN2 had the highest expression levels and was clearly not repressed in the subtropical environment.
Relative expression of three candidate genes for DCI and CQ. The expression of candidate genes residing in the identified QTL intervals, BoFLC3, PAN, and VRN2 were analyzed in four different tissues, the vegetative leaf at 4-leaf stage (VL), the latest leaf at curd forming stage (CL), the peduncle bract at harvest stage (PB), and floral bud at harvest stage (FB). Asterisks indicate significant difference at p < 0.05 between the two parents, BLM29 (closed box ■) and BLM25 (open box □), as analyzed by t test.
Association analysis of candidate genes for days to curd induction
To association between BoFLC3, PAN, and VRN2 and DCI, a panel of 95 broccoli-derived and 17 Chinese kale-derived breeding lines, were subjected to DCI measurement and haplotype analysis. After sequencing of the three genes in the 112 breeding lines, 3, 2, and 2 major haplotypes were identified for BoFLC3, PAN, and VRN2 (Fig. 2; Supplementary Fig. 4). Haplotypes of BoFLC3 and PAN, but not VRN2, accounted for DCI variation revealed by association analysis (Table 2). Among three major haplotypes of BoFLC3 (Fig. 2; Supplementary Fig. 4a), the DCI of haplotype G1 was significantly earlier than those of G2 and G3 in both 2014 and 2015 (Table 2) with high heritabilities (H2B = 0.9 in 2014 and 0.94 in 2015). Haplotype G1 shared the same sequence with the early parent line BLM29. Haplotype G2 exhibited a 13-bp insertion, a 255-bp deletion, and a 50-bp insertion at the first intron, a 37-bp insertion in intron 6, and 1 SNP at exons 2 and 4 leading to non-synonymous substitutions, K79N in an I-box domain and G110V in a K-box domain, respectively. Haplotype G3 shared nearly identical BoFLC3 sequence with G2 and the late parent, BLM25 except for a 13-bp insertion in intron 1 and a nonsynonymous substitution K151R on exon 6. The DCI of haplotype G1 was significantly earlier than those of G2 and G3 in both 2014 and 2015, indicating that the insertion and deletion on intron 1 and/or nonsynonymous substitutions of BoFLC3 altered DCI. Two major PAN haplotypes, G1 and G2, shared highly similar sequences with BLM29 and BLM25, respectively (Fig. 2, Supplementary Fig. 4b). The two PAN haplotypes, with high heritabilities (0.95 in 2014, 0.97 in 2015), had significant differences in average DCI in both years–G1 and G2 were 39.8 (±16.6) and 51.8 (±9.3) days in 2014, and 48.9 (±8.5) and 61.1 (±7.0) in 2015 (Table 2). Two VRN2 haplotypes shared high similarity with BLM29 and BLM25 (Fig. 2, Supplementary Fig. 4c), respectively, with low heritabilities (0.09 in 2014, 0.19 in 2015). The average DCI for haplotype G1 and G2 were 50.8 (±11.2) and 49.8 (±11.7) days in 2014, and 58.6 (±7.8) and 59.4 (±8.3) days in 2015 (Table 2), non-significant differences between haplotypes.
Albeit on different chromosomes, strong association was found between BoFLC3 and PAN haplotypes. Only 3 of 6 possible combinations of the three BoFLC3 and two PAN haplotypes occurred in the 112 accessions, BoFLC3-G1/PAN-G1, BoFLC3-G2/PAN-G2, and BoFLC3-G3/PAN-G2 (Table 3), deviating significantly from random association (χ2 = 111.2, p ≈ 0.001). The average DCIs of BoFLC3-G1/PAN-G1 were significantly earlier than BoFLC3-G2/PAN-G2, and BoFLC3-G3/PAN-G2, which were not significantly different from one another. No breeding lines carried early alleles at one locus and late alleles at the other, e.g., BoFLC3-G1/PAN-G2, BoFLC3-G2/PAN-G2 or BoFLC3-G3/PAN-G2 genotypes, indicating selection against such genotypes in high ambient temperature. Occurrence of the 6 possible BoFLC3/VRN2 genotype combinations slightly deviated from the random expectation (χ2 = 7.27, p ≈ 0.026), and DCI differences among these combinations were small (Table 3).
Discussion
Since the Brassicaceae originated in temperate zones, most flowering time mechanisms investigated to date regard long-day photoperiod and vernalization. In this study, 112 advanced broccoli breeding lines selected in subtropical regions provide a unique genetic resource to uncover new genes/QTLs. By bi-parental mapping, 4 DCI QTLs and 3 CQ QTLs accounted for totals of 56.28% and 33.02% of variance in curding time and curd quality (Table 1; Fig. 1). The location of qDCI-3/qCQ-3 corresponded to BoFLC313, and three BoFLC3 haplotypes were identified in these advanced breeding lines of broccoli (Fig. 2; Supplementary Fig. 4a). Two QTLs, qDCI-4 and qCQ-2 might coincide with QTLs of curd induction time uncovered in 111 cauliflower lines cultivated under cool circumstances but not high ambient temperature26. The other 2 QTLs, qDCI-6/qCQ-6 and qDCI-7 were newly uncovered in these two non-vernalized broccoli lines. Thus, we discovered novel identified genes/QTLs of DCI and CQ with new roles in floral regulation in relatively high ambient temperature under subtropical short-day conditions.
Vernalization, a long duration of low temperature, is a decisive environmental cue triggering flowering in Arabidopsis and some crops. B. oleracea crops can be classified by vernalization type as biennials (e. g. cabbage, kohlrabi) with non-vernalization types being annuals (e. g. broccoli and cauliflower)12. On the other hand, other reports indicate most broccoli cultivars require vernalization with a long duration below 23 °C4, but several commercial hybrid and breeding lines form curd at temperatures of 25–32 °C28,29. The present results indicated that DCI may be controlled by both vernalization and non-vernalization pathways.
Duplicated BoFLC genes permitted evolution of alternative pathways for control of flowering in temperate and tropical environments (Fig. 4). Gene redundancy contributes to diverse flowering time responses in Brassica15,16,17, however no reports to date reveal which of the four recognized FLC paralog(s) (BoFLC1, BoFLC2, BoFLC3, and BoFLC5) regulate flowering time in non-vernalizing types12,13,33. BoFLC2 explains large FTi variation in the F2 progeny of a broccoli (non-vernalization type)× cabbage (vernalization type) cross12, with expression reduced dramatically after vernalization, resulting in increased apex FT expression11,23. Null
Boflc2 alleles caused by a single base deletion in exon 4 are absent from biennials such as Brussels sprouts and cabbage but widespread in non-vernalizing broccoli, cauliflower, and a rapid cycling line12,13,34, including both parental lines BLM29 and BLM25 (Supplementary Fig. 5). It is possible that null function alleles of Boflc2 have been fixed in the annual Brassica crops, such as broccoli, cauliflower, and kale, since vernalization is unfavorable for cultivation of these crops in high temperature areas. In the DH lines of a rapid cycling line (var. alboglabra, non vernalization type) × a broccoli line (var. italica, non vernalization type), none of the 4 BoFLC were associated with FTi by linkage analysis, and BoFLC4 (BoFLC2) and BoFLC5 did not contribute to flowering time because of premature stop codons13. To date, no reports reveal which FLC paralog(s) play important role(s) in regulating flowering time in non-vernalization type plants.
BoFLC3, rather than BoFLC2, was the primary determinant of DCI variation in subtropical broccoli. We found no DCI QTLs near BoFLC2, but qDCI-3 and qCQ-3 overlapped one another and other flowering QTLs13,35, with BoFLC3 near the likelihood peak showing allelic variation between the parental lines. BLM29 and BLM25 shared the same amino acid sequences as the early flowering line A12DHd which is a rapid-cycling line derived from B. oleracea var. alboglabra and the late flowering line GDDH33 which is double haploid line derived from F1 hybrid Calabrese variety, Green Duke (B. oleracea var. italica)13, respectively. Both BLM29 and A12DHd alleles of BoFLC3 delayed FTi in the F2 and BC1S1 populations13, respectively (Table 1, Fig. 2). FLC2 dominated flowering time variation in B. oleracea and B. rapa populations responsive to vernalization, with no QTLs detected near FLC312,15. Moreover, BoFLC3 expressed consistently in cotyledons, leaves, and apex of broccoli and leaves of cauliflower and cabbage11,12. Thus, functional variation in BoFLC3 had little if any phenotypic effect under temperate environments. BLM29 had higher expression of BoFLC3 than BLM25 in VL, CL, and FL, which might cause earliness (Fig. 3). Thus, allelic effects and differential expression of BoFLC3 bring out various flowering times in natural broccoli germplasm. How BoFLC3 inhibits FTi by repressing FT and SOC1 remains of interest and its function is still to be determined.
The expression of FLC regulated by vernalization, especially the histone 3 modification on intron 1 by the activities of both VRN1 and VRN2, is well known and extensively investigated36. VRN2, corresponding to Bol032823, was a positional candidate gene for the major QTL on chromosome O6, qDCI-6/qCQ-6. VRN2 showed allelic variation in BLM29, BLM25, and cabbage (Fig. 2). Under the subtropical environment, expression levels of VRN2 were neither repressed nor significantly different between two parental lines (Fig. 3). Furthermore, genetic association between BoFLC3 and VRN2 was relatively small, a finding which might be caused by VRN2 being closely linked to the other real flowering gene(s) (Table 3). Together with lack of significantly different DCI between haplotypes, the results herein did not support that VRN2 regulated BoFLC3 to cause DCI variation in subtropical germplasm. BoVRN2/BoFLC2-independent mechanisms were suggested in temperature-regulated floral transition in cauliflower because of a lack of consistent relevance between gene functions of BoVRN2 and BoFLC226, and FLC-independent vernalization pathways were also suggested in broccoli and Arabidopsis37. Non-vernalization pathways are suggested to regulate DCI in subtropical germplasm, leading from obligate to facultative vernalization flowering.
Improved thermal tolerance of broccoli, reducing unfavorable characters such as leafy curds and uneven-sized flower buds3, was associated with qDCI-6/qCQ-6 here and a previously-published QTL22, for which PAN is the most likely candidate (Table 2; Fig. 2). The highest gene expression of PAN, a gene in which mutation changed shoot apical meristem size and floral organ number32, was detected in the FB of BLM25 (Fig. 3), perhaps contributing to good curd quality. One QTL on chromosome O6 induced curd formation when cauliflower was cultivated under temperatures >22 °C22. BoAP1-a was strongly suggested to control curd induction and bract development22,38; however, no amino acid variation was detected between BLM29 and BLM25 – indeed, only one SNP occurred even −139 bp upstream of BoAP1. Nevertheless, further investigations are needed to prove the role of PAN.
The wide range of flowering times in natural population is regulated not only by multiple genes in accordance to environmental cues but also by intrinsic non-linear gene interactions (epistasis). The most well-known of these interactions is that FLC gene expression is repressed by VRN2 and VIN3, and consequently the expression of FT and SOC is activated10,36. Digenic interaction contributed 3.2% heat tolerance variation in a broccoli DH population39. In the BLM29 × BLM25 F2 population, two digenic interactions accounted for DCI but not CQ variation, in which qDCI-6 interacted with qDCI-3 and qDCI-7, respectively (Table 1). Thus, the novel QTL, qDCI-6, played an important role in regulating FTi by interacting with other genes. The candidate gene for qDCI-6, PAN, was strongly associated with BoFLC3 in 112 subtropical broccoli breeding lines (Table 3), strongly suggesting that specific combinations of BoFLC3 and PAN haplotypes confer selective advantages under relatively high ambient temperature and short-day length.
The adjustment of crop flowering time has been a breeding goal of widespread importance to maximize harvestable yields and shift cultivation seasons to meet market demands. To promote broccoli to form curd too early in Southeast Asia would risk heat stress, resulting in various-sized floral buds, poor curd quality with several bracts of leaves and perhaps even failure of curd initiation (Fig. 1a). Alleles conferring earliness and good curd quality are both necessary to commercially successful elite cultivars. In this study, BLM29 provided the favorable allele, promoting curding by 3.81 days and reducing curd leafiness by 0.44 degree, from qDCI-6/qCQ-6 (Table 1). After 3 backcross generations with marker-assisted selection, four selected BC3F1 individuals exhibited earlier flowering time than the elite parent line BLM25 but possesses similarly superior horticultural traits (Supplementary Figure 3). In this study, the novel FTi QTL, qDCI-6/qCQ-6 conferring curd induction time and curd quality provides a valuable genetic resource for breeding broccoli, cauliflower and other Brassica vegetable cultivars optimized for yield and quality in the climatic zones of relatively high ambient temperature.
Evolution of alternative pathways for control of flowering in temperate and tropical environments respectively, based on duplicated genes, might be utilized in adaptation of numerous taxa to new climates, either by searching for naturally occurring gene duplicates or using gene engineering and/or editing approaches. As ambient temperatures rise, plant adaptation to diverse geography and maximum crop production is likely to involve extensive modification of temperature responses, as exemplified by the adaptation of B. oleracea to Taiwan. Indeed, reconciling current models for the genetics of flowering with results from divergent latitudes or warmer climates requires new dimensions (Fig. 4). Linearity and additivity may be inadequate in such models – the wide range of flowering times in natural populations is regulated not only by individual gene effects but also by non-linear interactions (epistasis) such as qDCI-6 with qDCI-3 and qDCI-7, respectively (Table 1). This study provided new insight into FTi regulation and implemented a strategy to breed new elite lines with earliness, demonstrating an example of translational agriculture.
Methods
Plant material
To identify QTLs conferring curd induction time and curd quality in broccoli, B. oleracea var. italica, an F2 population derived from a cross between two commercial breeding lines, BLM25 and BLM29, was established. BLM25, with superior curd quality and horticultural characters but with late curding time was used as a female parent in the F1 cross and as the recurrent parent in subsequent marker-assisted backcrossing. BLM29, a kale-derived broccoli breeding line with early curd formation but inferior curd quality was the male parent and donor parent. The randomly selected 94 and 188 F2 individuals were used to construct a primary linkage map and map QTLs conferring two important traits, days to induction (DCI) and curd quality (CQ). The 188 F2 individuals were cultivated in conventional cool seasons starting from Oct 1, 2010 to Jan 14, 2011 to evaluate DCI, and their derived F2.3 families were grown in the hot season starting from July 12 and initiated curd formation about September 10, 2011 to evaluate CQ.
A panel of 112 advanced broccoli breeding lines were genotyped to identify haplotypes of candidate genes for association genetic analysis, and phenotyped in the fall-winter of 2014 and 2015. All plants were grown at the experimental station of Known-You Seed Co., Ltd, Xinshi, Tainan, Taiwan (23.079337°N, 120.295377°E).
Assessment of days to induction and curd quality
Days to induction (DCI) for each F2 individual was determined by the duration from the day of seedling transplantation to the field until curd initiation, indicated by visible curd of approximately 0.5 cm diameter. Curd quality (CQ) of each F2 individual was estimated by the arithmetic mean of 30 F2:3 individuals. CQ, reflected by curd shape, was evaluated based on curd leafiness reflected by abundance of bracts appearing between inflorescence internodes at harvest. CQ index was scored from 1 to 4 according to the abundance of bracts, from none, few, several, to numerous bracts (Fig. 1).
Genotyping assay of molecular markers
Genomic DNA extraction and genotype assays of PCR-based markers were as described previously40,41. SSR markers with prefixes of BnGMS, Ol, BoGMS, fito, BRMS, Na, CB, BRAS, SORA, and BrGMS were retrieved from previous publications and described well in Shuang et al.40 and intron polymorphic (IP) markers with prefix At were adopted from Panjabi et al.42. After genotype assay, 117 of 402 SSR markers and 61 of 100 IP markers exhibited polymorphism between BLM25 and BLM29. To fill linkage gaps, SSRs with prefix of CHT were mined from a whole genome shotgun sequence of BLM29 obtained by 454 (Roche), and the primer information was listed as in Supplementary Table 4.
Genetic mapping of QTLs conferring curd induction time and curd quality
The primary linkage map was constructed by using 94 randomly selected F2 progeny genotyped with 126 polymorphic markers, including 107 SSR and 19 IP markers distributed on the 9 broccoli chromosomes (Supplementary Figure 1). For interval mapping of QTLs, a total of 188 F2 individuals were genotyped with 60 markers at average spacing of approximately 15 cM. The linkage map was constructed by using MapMaker Exp 3.0 with a LOD threshold of 3.543. To identify QTLs of DCI and CQ, a multiple-QTL mapping analysis based on Haley-Knott regression was performed using R/qtl44. Forward QTL mapping analysis was applied, with a single-QTL genome scan performed first and followed with incorporation of QTL with the largest effect into the model by functions scanone and makeqtl. Additional additive QTL and interaction pairs were then scanned and evaluated in the model by functions scantwo, makeqtl, and addint. The map position of each QTL in the model was refined by function refineqtl. The whole procedure was repeated until no more significant QTL was detected.
Breeding elite early curding lines by marker-assisted selection
The scheme for breeding elite early curding lines by marker-assisted selection (MAS) was illustrated on Supplementary Figure 2. The 188 F2 individuals used for QTL analysis were self-pollinated to generate 188 F2:3 families which were genotyped with fito203 and fito036, flanking qDCI-6, for foreground selection and 22 markers for background selection. Three F2:3 were selected and backcrossed to the elite parent, BLM25, to generate the BC1F1 population. After MAS with 50 markers, 10 of 300 BC1F1 individuals were subsequently backcrossed to BLM25 to generate 192 BC2F1 individuals. After MAS, 4 selected BC2F1 individuals were self-pollinated several times to generate 128 BC2F2 individuals and backcrossed to BLM25 to generate 128 BC3F1 individuals. Four of 128 BC3F1 individuals were selected and were consequently self-pollinated several times to generate 90 BC3F2 individuals.
Only 113 BC2F2 and 87 BC3F2 individuals were grown from February 15 and October 5, 2015 respectively, to evaluate DCI and genotyped with fito203 and fito036 to predict the genotypes of qDCI-6. Statistical summary parameters, one-way analysis of variance, and Fisher’s least significant difference (LSD) were calculated by using R45.
Identification of candidate genes for curd induction and curd quality
The whole genome sequence of B. oleracea, assembly GCA_000695525.1, was obtained from EnsemblPlants (http://plants.ensembl. org/index.html). Gene annotation was performed for 2 major QTL intervals, At5g07910/CHT20 on chromosome O3 and fito203/fito036 on chromosome O6. Genes with putative functions in flowering pathways and floral organ differentiation were considered candidates for DCI and CQ.
Sequences of candidate genes were downloaded from a Brassica database (http://brassicadb.org/). Candidate genes were sequenced from approximately 1,000 bp upstream of the transcription start site to 100–300 bp downstream of the transcription stop codon. In addition to sequencing the candidate genes residing in the two major QTL intervals, one important flowering time gene, BoFLC2, was genotyped according to Ridge et al.34.
Quantitative real-time PCR analysis of the expression of candidate genes
RNA from BLM29 and BLM25 was extracted from approximately 1 g of four tissues at the vegetative leaf at 4-leaf stage (VL), the latest leaf at curd forming stage (CL), the peduncle bract at harvest stage (PB), and floral bud at harvest stage (FB) by using TRIzol Reagent (Invitrogen, USA). The first-strand cDNA was synthesized with 1 μg total RNA using PrimeScript™ RT Reagent Kit (Perfect Real Time, TAKARA Bio Inc., Japan) in a volume of 10 μL, and qPCR was carried out using an ABI 7500 Sequence Detection System (ABIPRISM; Applied Biosystems, USA) with the KAPA SYBR® FAST qPCR Kits (KAPA Biosystems, USA) in a total volume of 20 μL. The relative expression of genes was calculated by using BoActin as the internal control.
Association analysis of DCI candidate genes
To identify the most likely DCI candidate genes, DCI of 112 commercial broccoli breeding lines were estimated and candidate genes sequenced. The broccoli was grown in Xinshi Dist., Tainan, in the conventional cool seasons starting from September 2014 and 2015, respectively. DCI of each line was estimated from the average of 8–10 individuals. Broad sense heritability was calculated as mean square of genotype divided by total mean square (H2B = VG/VP). The haplotypes of breeding lines were identified according to major DNA variants of the candidate genes.
References
Warwick, S. I. & Black, L. D. Molecular systematics of Brassica and allied genera (Subtribe Brassicinae, Brassiceae) - chloroplast genome and cytodeme congruence. Theor. Appl. Genet. 82, 81–92 (1991).
Kushad, M. M. et al. Variation of glucosinolates in vegetable crops of Brassica oleracea. J. Agric. Food Chem. 47, 1541–1548 (1999).
Björkman, T. & Pearson, K. J. High temperature arrest of inflorescence development in broccoli (Brassica oleracea var. italica L.). J. Exp. Bot. 49, 101–106 (1998).
Wurr, D., Fellows, J., Phelps, K. & Reader, R. Vernalization in calabrese (Brassica oleracea var. italica)-a model for apex development. J. Exp. Bot. 46, 1487–1496 (1995).
Jung, C. & Müller, A. E. Flowering time control and applications in plant breeding. Trends Plant Sci. 14, 563–573 (2009).
Kazan, K. & Lyons, R. The link between flowering time and stress tolerance. J. Exp. Bot. 67, 47–60 (2016).
Bäurle, I. & Dean, C. The timing of developmental transitions in plants. Cell 125, 655–664 (2006).
Srikanth, A. & Schmid, M. Regulation of flowering time: all roads lead to Rome. Cell Mol. Life Sci 68, 2013–2037 (2011).
Bouché, F., Lobet, G., Tocquin, P. & Périlleux, C. FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res. 44, D1167–71 (2016).
Michaels, S. D. & Amasino, R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Lin, S.-I. et al. Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol. 137, 1037–1048 (2005).
Okazaki, K. et al. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor. Appl. Genet. 114, 595–608 (2007).
Razi, H., Howell, E., Newbury, H. & Kearsey, M. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor. Appl. Genet. 116, 179–192 (2008).
Liu, S. et al. The Brassica oleraceae genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930 (2014).
Schranz, M. E. et al. Characterization and effects of the replicated flowering time gene FLC In Brassica rapa. Genetics 162, 1457–1468 (2002).
Tadege, M. et al. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 28, 545–553 (2001).
Golicz, A. A. et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 7, 13390 (2016).
Schiessl, S. V., Huettel, B., Kuehn, D., Reinhardt, R. & Snowdon, R. J. Flowering time gene variation in Brassica species shows evolutionary principles. Front. Plant Sci. 8, 1742 (2017).
Kowalski, S. D., Lan, T.-H., Feldmann, K. A. & Paterson, A. H. Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved gene organization. Genetics 138, 499–510 (1994).
Li, G., Gao, M., Yang, B. & Quiros, C. Gene for gene alignment between the Brassica and Arabidopsis genomes by direct transcriptome mapping. Theor. Appl. Genet. 107, 168–180 (2003).
Axeisson, T., Shavorskaya, O. & Lagercrantz, U. Multiple flowering time QTLs within several Brassica species could be the result of duplicated copies of one ancestral gene. Genome 44, 856–864 (2001).
Hasan, Y. et al. Quantitative trait loci controlling leaf appearance and curd initiation of cauliflower in relation to temperature. Theor. Appl. Genet. 129, 1273–1288 (2016).
Irwin, J. A. et al. Nucleotide polymorphism affecting FLC expression underpins curding date variation in horticultural brassicas. Plant J. 87, 597–605 (2016).
Lan, T.-H. & Paterson, A. H. Comparative evolution of quantitative trait loci sculpting the curd of Brassica oleracea. Genetics 155, 1927–1954 (2000).
Lan, T. H. & Paterson, A. H. Comparative mapping of QTLs determining the plant size of Brassica oleracea. Theor. Appl. Genet. 103, 383–397 (2001).
Matschegewski, C. et al. Genetic variation of temperature-regulated curd induction in cauliflower: elucidation of floral transition by genome-wide association mapping and gene expression analysis. Front. Plant Sci. 6, 720 (2015).
Zhao, J. et al. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J. Exp. Bot. 61, 1817–1825 (2010).
Yang, Y. W., Tsai, C. C. & Wang, T. T. A heat-tolerant broccoli F1 hybrid, ‘Ching-Long 45’. HortSci. 33, 1090–1091 (1998).
Farnham, M. W. & Björkman, T. Evaluation of experimental broccoli hybrids developed for summer production in the eastern United States. HortSci. 46, 858–863 (2011).
Duclos, D. V. & Björkman, T. Meristem identity gene expression during curd proliferation and flower initiation in Brassica oleracea. J. Exp. Bot. 59, 421–433 (2008).
Sotelo, T., Soengas, P., Velasco, P., Rodríguez, V. M. & Cartea, M. E. Identification of metabolic QTLs and candidate genes for glucosinolate synthesis in Brassica oleracea leaves, seeds and flower buds. PLoS One 9, e91428 (2014).
Maier, A. T., Stehling-Sun, S., Offenburger, S. L. & Lohmann, J. U. The bZIP transcription factor PERIANTHIA: a multifunctional hub for meristem control. Front. Plant Sci. 2, 79 (2011).
Simpson, G. G. & Dean, C. Arabidopsis, the Rosetta stone of flowering time? Science 296, 285–289 (2002).
Ridge, S., Brown, P. H., Hecht, V., Driessen, R. G. & Weller, J. L. The role of BoFLC2 in cauliflower (Brassica oleracea var. botrytis L.) reproductive development. J. Exp. Bot. 66, 125–135 (2015).
Uptmoor, R., Schrag, T., Stützel, H. & Esch, E. Crop model based QTL analysis across environments and QTL based estimation of time to floral induction and flowering in Brassica oleracea. Mol. Breeding 21, 205–216 (2008).
Sheldon, C. C., Finnegan, E. J., Dennis, E. S. & Peacock, W. J. Quantitative effects of vernalization on FLC and SOC1 expression. Plant J. 45, 871–883 (2006).
Uptmoor, R., Li, J., Schrag, T. & Stützel, H. Prediction of flowering time in Brassica oleracea using a quantitative trait loci-based phenology model. Plant Biol. 14, 179–89 (2012).
Labate, A., Robertson, L. D., Baldo, A. M. & Björkman, T. Inflorescence identity gene alleles are poor predictors of inflorescence type in broccoli and cauliflower. J. Amer. Soc. Flort. Su. 131, 667–673 (2006).
Branham, S. E., Stansell, Z. J., Couillard, D. M. & Farnham, M. W. Quantitative trait loci mapping of heat tolerance in broccoli (Brassica oleracea var. italica) using genotyping-by-sequencing. Theor. Appl. Genet. 130, 529–538 (2017).
Shuang, L.-S. et al. Application of Molecular markers of Brassicaceae in the polymorphic and phylogenetic analyses of broccoli and cauliflower. J. Taiwan Soc. Hort. Sci. 58, 45–60 (2012).
Hsu, Y. C. et al. Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol Breed. 34, 655–673 (2014).
Panjabi, P. et al. Comparative mapping of Brassica juncea and Arabidopsis thaliana using Intron Polymorphism (IP) markers: homoeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9, 113 (2008).
Lander, E. S. et al. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181 (1987).
Arends, D., Prins, P., Jansen, R. C. & Broman, K. W. R/qtl: high-throughput multiple QTL mapping. Bioinformatics 26, 2990–2992 (2010).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2013).
Chow, C.-N. et al. PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, D1154–D1160 (2016).
Acknowledgements
This work was funded by the Council of Agriculture of Taiwan (99AS-5.3.1-ST-a8, 102AS-6.2.1-ST-a2) to K.H. and the Ministry of Science and Technology (MOST 103-2321-B-002-067, MOST 104-2321-B-002-063) of Taiwan to Y.L. The authors are grateful to Yu-feng Chan and Yao-chuan Yu for their contribution to data collection.
Author information
Authors and Affiliations
Contributions
Y.L. and K.H. designed and supervised the research. C.L. and C.S. contributed to prepare genetic materials, conduct field trails and collect phenotype data. M.T., J.L., C.W., C.L. and L.S. conducted genotype and sequence assays and analyzed data. Y.L., M.T. and J.L. wrote the manuscript. Y.L., A.P. and K.H. edited and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, Yr., Lee, Jy., Tseng, Mc. et al. Subtropical adaptation of a temperate plant (Brassica oleracea var. italica) utilizes non-vernalization-responsive QTLs. Sci Rep 8, 13609 (2018). https://doi.org/10.1038/s41598-018-31987-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-31987-1
Keywords
This article is cited by
-
Upregulation of tandem duplicated BoFLC1 genes is associated with the non-flowering trait in Brassica oleracea var. capitata
Theoretical and Applied Genetics (2023)
-
A 215-bp indel at intron I of BoFLC2 affects flowering time in Brassica oleracea var. capitata during vernalization
Theoretical and Applied Genetics (2022)
-
Genome sequencing sheds light on the contribution of structural variants to Brassica oleracea diversification
BMC Biology (2021)
-
Understanding population structure and detection of QTLs for curding-related traits in Indian cauliflower by genotyping by sequencing analysis
Functional & Integrative Genomics (2021)
-
Current understanding of flowering pathways in plants: focusing on the vernalization pathway in Arabidopsis and several vegetable crop plants
Horticulture, Environment, and Biotechnology (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.