Abstract
Osteosarcoma (OS) is the most common type of primary malignant bone tumor and mainly occurs in children and adolescent. Because of its early migration and invasion, OS has a poor prognosis. It has been reported that mircoRNAs (miRNAs) play a crucial role in the occurrence and development of multiple tumors. In this study, we identified the aberrant-expression of miR-143-3p in osteosarcoma and examined the role of miR-143-3p in OS development. Further, we searched the miR-143-3p target gene and verified its accuracy by luciferase experiments. Finally, we explored the relationship between miR-143-3p and FOS-Like antigen 2 (FOSL2). Our data indicated that miR-143-3p expression was substantially lower in OS tissues and cell-line compared with normal tissues, and was lower in patients with poor prognosis. In addition miR-143-3p inhibited OS cell proliferation and metastasis while promoting apoptosis. We next showed that FOSL2 was directly targeted by miR-143-3p and could reverse the inhibition caused by miR-143-3p. Finally, we found FOSL2 expression in OS cells was significantly higher compared with normal cells and negatively correlated with miR-143-3p. Thus, miR-143-3p directly and negatively targets FOSL2 to affect OS characteristics. This provides a new target for the treatment of OS and deserves further study.
Similar content being viewed by others
Introduction
Osteosarcoma (OS) is a common primary malignant bone tumor occurring in adolescents and children especially in those under 20 years of age1. At present, the origin of OS is not clear. Some reports suggest that OS may originate from mesenchymal cells2,3. With its characteristic early metastasis, the prognosis of OS is usually poor. The tumor cells can break through the cortex and medullary cavity and transfer through the blood to other tissues, especially the lung4,5. A small number of tumor cells can transfer to the brain, prostate and other tissues and lead to death6. Although, in recent years, the 5-year-survival-rate of OS patients has shown some increase with the improvement of medical technology, the survival rate is still very low3. At present, the metastasis of OS is still the main factor affecting the prognosis of OS patients.
MicroRNA (miRNA) is a single-stranded non-coding RNA containing 22–24 nucleotides and exists on non-coding regions7. As a regulator of mRNA8, it regulates the entire process of cell proliferation, gene expression and even ontogenesis9. It has been reported that abnormal expression of miRNAs is found in a large number of human tumors including renal carcinoma, colon cancer, lung cancer, glioblastoma and gastric cancer6,10,11. Recent studies have revealed that many miRNAs are expressed abnormally in OS cells and play a role as oncogenes or anti-oncogenes in the occurrence and development of OS12,13.
In this study, we examined differences in miR-143-3p expression between and normal tissues and explored the biological function of miR143-3p in proliferation, apoptosis, migration and invasion in OS cell lines and tissues. At the same time, we investigated the relationship between miR-143-3p and its target gene FOS-like antigen 2 (FOSL2) in the regulation of OS.
Results
miR-143-3p is significantly downregulated in OS tissues and OS cell lines
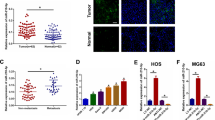
We collected OS tissues and adjacent normal tissues from 20 tumor cases, and measured the expression of miR-143-3p in each specimen. qRT-PCR analysis showed that the expression of miR-143-3p was significantly lower in OS cells than in normal tissues (Fig. 1A). The ratio of miR-143-3p expression in normal tissue and tumor tissue was greater than 2 (N/T > 2) in 14 of 20 (70%) patients (Fig. 1B). At the same time, we analyzed the expression of miR-143-3p in three cell lines: MG-63, 143B and the normal human osteoblastic cell line hFOB 1.19, and found that the expression level of miR-143-3p in the human OS cell lines (MG-63, 143B) was significantly lower than that in normal human osteoblastic cells (Fig. 1C). The analysis of the relationship between miR-143-3p expression level and clinical features of OS patients is shown in Table 1.
miR-143-3p downregulation in OS tissues and cell lines is associated with poor prognosis. (A) The miR-143-3p expression is significantly decreased in OS tissues. (B) miR-143-3p expression relative to adjacent normal tissues for 20 patients. (C) The miR-143-3p expression in MG-63 and 143B cell lines are significantly lower compare to hFOB 1.19 cell line determined by qRT-PCR. (D) Kaplan-Meier curves were performed for survival in all 33 patients.
miR-143-3p expression is lower in poor prognosis patients
To determine whether miR-143-3p expression correlates with clinical prognosis in patients, we gathered information of all 33 OS patients and used RT-PCR to measure the expression of miR-143-3p. The median value of miR-143-3p expression in all patients was used to divide the patients into two groups: a high expression group (>median, n = 10) and low expression group (<median, n = 23). From these statistical results, we found that the survival time of the high-expression group was longer compared to the low expression group (Fig. 1D).
miR-143-3p inhibits proliferation of OS cells
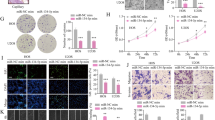
To determine the effect of miR-143-3p on proliferation of OS cells, we transfected miR-NC, miR-143-3p mimic, miR-NC-inhibitor and miR-143-3p inhibitor into the two cell lines. The CCK-8 assay was used to measure cell proliferation. The results showed that the proliferation of MG-63 and 143B cells after transfection with miR-143-3p mimic was significantly inhibited compared with the miR-NC group. Meanwhile transfection of the miR-NC-inhibitor and miR-143-3p inhibitor significantly increased the proliferation of MG-63 and 143B cell lines compared to the miR-NC-inhibitor group (Fig. 2A). These results indicated that miR-143-3p could inhibit the growth of OS cells. The results of cell cycle analysis showed that up-regulation of miR143-3p increases the number of cells in G1 phase and synchronously decreases the number in S phase, whereas inhibition of miR-143-3p decreases the number of cells in G1 phase and increases the number in S phase (Fig. 2B and C). All these results indicated that miR-143-3p could inhibit proliferation by blocking the G1/S phase transition. Moreover, the apoptosis rate was significantly higher in the two cell lines over-expressing miR-143-3p (Fig. 2D).
miR-143-3p inhibits proliferation of OS cells. (A) MG-63 and 143B cell lines were transfected with miR-143-3p mimics and miR-NC and the cell viability determined with the CCK-8 assay. (B) The results of flow cytometry analysis of MG-63 and 143B cell lines after transfected with miR-144 mimics and miR-NC. (C) The results of flow cytometry analysis of 143B cell lines after transfected with miR-143-3p-in and NC-in. (D) Results of colony formation and apoptosis cells of MG-63and 143B cell lines transfected with miR-143-3p mimics and miR-NC.
miR-143-3p expression inhibits migration and invasion of OS cells in vitro
To further explore the function of miR-143-3p in migration and invasion of OS, we transfected the miR-NC, miR-143-3p mimic, miR-NC-inhibitor and miR-143-3p inhibitor into MG-63 and 143B cell lines. The wound healing assay showed that after up-regulation of miR-143-3p expression, migration and invasion of tumor cells was decreased compared with the control group (Fig. 3A and B). In contrast down-regulating miR-143-3p expression would lead to increased migration compared with the control group (Fig. 3C and D). The invasion assay showed that the invasive ability of the two cell lines was decreased in the miR-143-3p mimic group compared to the control group, while inhibiting miR-143-3p expression increased the cell’s invasive ability compared to the control group (Fig. 3E and F). This shows that miR-143-3p can inhibit cell migration in vitro.
miR-143-3p expression inhibits migration and invasion of OS cells in vitro. (A,B) Wound-closure assay for MG-63 and 143B cell lines transfected with miR-144 mimics and miR-NC. (C,D) Wound-closure assay for MG-63 and 143B cell lines transfected with miR-143-3p-in and NC-in. (E) Images of the Transwell invasion assay of MG-63 cell line transfected with miR-144 mimics, miR-NC, miR-143-3p-in and NC-in. (F) Quantification of Transwell invasion assay for MG-63 and143B cell lines.
miR-143-3p inhibit tumor growth and metastasis in vivo
Based on the foregoing results, we conducted animal experiments to determine the function of miR-143-3p in vivo. The two cell lines which could stably express the miR-143-3p mimic and miR-NC were injected subcutaneously into nude mice and the animals were monitored for tumor growth for 4 weeks. The results showed that tumors which expressed miR-143-3p mimic were significantly smaller in volume and weight compared to the control group (Fig. 4A and B). Lung tissue of nude mice was also removed to determine the effects of miR-143-3p on tumor metastasis. We found that the number of lung metastasis nodules was dramatically decreased in the miR-143-3p group compared with the control group (Fig. 4C). This indicates that miR-143-3p could inhibit tumor growth and metastasis in vivo. Another group of mice was monitored for 100 days to construct a survival curve. The results showed that the survival time and rate of mice in high miR143-3p expression group were significantly higher than in the control group (Fig. 4D).
miR-143-3p inhibit tumor growth and metastasis in vivo. (A) Photographs of tumors (B) Curve of tumor volume growth for the nude mice. (C) Tumor of weight. (D) KI-67 staining section of lung tissue. (E) Number of lung metastatic nodules of each group. (F) Survival curve of the two groups mice injected the cell line respectively which could stably express the miR-143-3p mimics and miR-NC.
miR-143-3p directly targets FOSL2
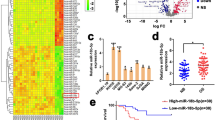
We used the TargetScan, MiRanda, and PicTar databases to retrieve and draw the result of the search and identified FOSL2 as a possible target gene for miR-143-3p (Fig. 5A). To further verify its accuracy, we transfected the miR-143-3p mimic and miR-NC into MG-63 and 143B cell lines. Through this experiment we found that increasing the expression of miR-143-3p in cells significantly decreased the expression of FOSL2 compared to the miR-NC group (Fig. 5B and C). Then we transfected the miR-143-3p inhibitor and miR-NC-inhibitor into the two cell lines. The results showed that the expression of FOSL2 significantly increased compared to the control group (Fig. 5B and D). A luciferase reporter gene test showed that miR-143-3p mimic significantly decreased the activity of FOSL2 3′-UTR compared with the mutant group (Fig. 5E). Spearman’s correlation analysis was carried out using SPSS 22.0 software (Fig. 5F). These data suggest that FOSL2 is the target gene of miR-143-3p and negatively correlated with miR-143-3p.
miR-143-3p directly targets FOSL2. (A) The sequences of the putative miR-143-3p binding sites in wild type and mutant (red) FOSL2-3′UTR. (B) Determination of FOSL2 protein in MG-63 cell after transfection with miR-144 mimics, inhibitors and miR-NC, NC-in or miR-144 lentivirus infection. (C) Determination of FOSL2 mRNA in MG-63 and 143B cell lines after transfected with miR-144 mimics and miR-NC. (D) FOSL2 mRNA in MG-63 and 143B cell lines after transfected with miR-143-3p-in and NC-in. (E) The relative luciferase activity of luciferase reports with wild type or mutant FOSL2-3′UTR were determined in MG-63 cell line, which were transfected with the miR-143-3p mimics or miR-NC. Statistical significance was observed between the wild type and the mutant groups. (F) Spearman’s correlation analysis between miR-143-3p expression and FOSL2 mRNA level.
FOSL2 downregulation is a critical step in regulation of OS properties by miR-143-3p
Based on the foregoing results, we speculated whether miR-143-3p affected OS characteristics through regulating FOSL2 expression. To verify our conjecture, we transfected construct containing FOSL2 without the 3′-UTR into the cell lines which could stably express the miR-143-3p mimic (Fig. 6A and B). The results showed that the ectopic expression of FOSL2 could significant reversed the inhibition caused by miR-143-3p. Meanwhile silencing FOSL2 by specific siRNA led to enhanced the inhibition caused by miR-143-3p (Fig. 6C–E). Therefore, FOSL2 down-regulation is a critical step in regulation of OS properties by miR-143-3p.
FOSL2 downregulation is a critical step in regulation of OS properties by miR-143-3p. (A) The miR-143-3p expression after transfected with lentivirus vectors. (B) (left) MG-63-miR cell (stably express miR-143-3p) were transfected with empty pcDNA3.1 vector, FOSL2-containing plasmid; (right) MG-63-miR cell were transfected with siRNA against FOSL2. (C) Cell viability of the two cell lines were measured by CCK-8 assay. The results showed that cell viability of ectopic FOSL2 group was higher than the control group. And the cell viability of siFOSL2 group was lower than NC group. (D) Cell cycle distribution were measured by flow cytometry analysis. The effects of G1/S phase blocking was weakened by ectopic FOSL2 and enhanced by siFOSL2. (E) Image and quantification of cell invasion ability were measured by Transwell assay.
Discussion
OS is a common malignant tumor of bone often occurring in children and adolescents under the age of 20 years14. Some reports suggest that OS may originate from mesenchymal cells as it is prone to local invasion and easily undergoes distant metastasis resulting in a poor prognosis and high mortality15. The metastasis of OS is always one of the main factors that influences the survival rate of OS patients3,16. Under current treatment, the 5-year-survival-rate of patients without metastasis is 60–70%, while the 3-year-survival-rate for patients with distant metastasis is less than 30%17,18. In addition, the lung is the main metastasis site of OS and lung metastasis is the most important and frequent cause of death19,20,21. Recent studies have found that multiple miRNAs are expressed abnormally in OS cells and play roles as oncogenes and tumor suppressor genes in the occurrence and development of OS10,12. As the related literature reports, miRNAs participate widely in tumor development and inhibit tumor occurrence and development through there effect on target genes in tumor; for example miR-143-3p inhibits prostate cancer invasion and metastasis by targeting gene MMP1322,23, and miR-497 and miR-195 inhibit the invasion and metastasis of breast cancer by targeting the protein Raf-1 and Ccnd124,25.
The down-regulation of miR-143 expression has also been reported in several human cancers, including colorectal cancer23, prostate cancer26, cervical cancer27, ovarian cancer28 and B-cell lymphoma29. Thus, it is considered that miR-143 is a tumor-suppressor miRNA. We analyzed expression of miR-143-3p in OS cells and adjacent normal tissues and found that the expression of miR-143-3p in tumor tissues decreased significantly compared with adjacent normal tissues, similar to previous report. This indicates that the decrease of miR-143-3p expression may play a role in the occurrence and development of OS. Low expression of miR-143-3p was linked to distant metastasis and poor prognosis in OS patients. This may provide a new way to predict the prognosis for OS patients.
In general, miRNAs play a regulatory role by directly inhibiting target genes30,31,32. To further study the mechanism of miR-143-3p in the occurrence and development of OS, we infected MG-63 and 143B cell lines using a specific virus. The effect of miR-143-3p on cell proliferation and apoptosis is controversial. Fan et al. reported that miR-143-3p could inhibit tumor cell proliferation and promote apoptosis23. Meanwhile Osaki et al. showed that miR-143-3p has no effect on the proliferation or apoptosis of OS33. Akao et al. reported that the target gene of miR-143 was determined to be ERK5/MAPK7, the up-regulation of which leads to cell growth via activation of c-Myc, in Raji cells, a human B-cell lymphoma cell line34. Recently, another publication showed that miR-143 suppressed cell proliferation by inhibiting KRAS translation in human colorectal cancer35,36. Our study found that the apoptosis rate of tumor cells increased significantly after up-regulation of miR-143-3p expression, indicating that miR-143-3p is involved in the process of apoptosis. Thus the decreased expression of miR-143-3p may lead to the development of tumors.
Cell migration experiments showed that in the MG-63 and 143B cell lines, cell migration ability increased significantly after knockdown of miR-143-3p expression, while expression increased after inhibition of miR-143-3p function. All the results showed that miR-143-3p was closely related with OS which was prone to migratory and invasive properties. These findings provide a new theoretical basis for further treatment of OS. Not only that, we can also diagnose and predict OS by detecting the expression of miR-143-3p in tissues, serum or other specimens.
The results of in vitro experiments showed that miR-143-3p-mimics inhibited the invasion and metastasis of OS cell lines. To investigate the detailed mechanism, we identified the target gene of miR-143-3p through searching TargetScan, MiRanda and PicTar databases and confirming the results using the Luciferase reporter gene test. FOSL2 belongs to the activator protein 1 (activator protein, l, APl) transcription factor family37,38. FOSL2 is widespread in human tissues and plays an important role in mammalian growth, reproductive traits, and immune response39,40. The expression products of FOSL2 are related to cell adhesion, movement, invasion, metastasis and cell growth37. Studies have shown that over-expression of FOSL2 is associated with a higher invasive association with breast cancer and involved in breast cancer progression39. In addition, FOSL2 also plays a role in the TGF- beta signaling pathway, and is essential for TGF- beta1 to induce cell migration.
Finally, our results showed that when the expression level of miR-143-3p in cells was increased, the expression level of FOSL2 decreased significantly. In contrast, inhibiting miR-143-3p expression led to a dramatic increase in the expression level of FOSL2. We also found that FOSL2 expression could significantiy reverse the inhibition caused by miR-143-3p. Furthermore, silencing of FOSL2 by specific siRNA enhanced the inhibitory effects caused by miR-143-3p. These data suggest that down-regulation of FOSL2 is a critical step in regulation of OS properties and the expression of miR-143-3p can inhibit the proliferation, migration and invasion of OS by reducing the expression of FOSL2 (Fig. 7).
In conclusion, the decrease of miR-143-3p expression in OS cells is closely related to the occurrence and development of OS. We have shown that miR-143-3p directly targets FOSL2 and affects the OS characteristics through down-regulation of FOSL2 expression. This provides us with a new target for the treatment of OS, and is deserving of further study.
Materials and Methods
OS tissue samples
The study was approved by the Ethics Committee of Renmin Hospital of Wuhan University. Written informed consent was obtained from the patients and their parents/guardians prior to tissue collection. All procedures were conducted in accordance with guidelines provided by the Ethics Committees and Institutional Review Boards. In brief, osteosarcoma tissue specimens (n = 33) were gathered from 2013 to 2017 at Renmin Hospital of Wuhan University and 20 adjacent normal tissues were collected at the time of operation simultaneously. The relevant clinical information was collected from patients with prior informed written consent. All osteosarcoma specimens were frozen in liquid nitrogen and then cryopreserved in −80 °C. Tumor stage and grade were classified according to the Enneking staging system.
Cell culture
Three cell lines: hFOB 1.19 (human fetal osteoblastic cell line), MG-63 and 143B were purchased from the Institute of Cell Bank for Biological Sciences (Shanghai, China). MG-63 and 143B were cultured in DMEM (Hyclone, Utah, USA) containing 10% FBS (Gibco, Australia) and 1% antibiotics (penicillin 100 U/ml, streptomycin 100 μg/ml, Hyclone, Utah, USA) at 37 °C with 5% CO2 and 100% humidity. hFOB1.19 cell line was maintained in DMEM/F-12 (Hyclone, Utah, USA) supplement 10% FBS (Gibco, Australia) and geneticin (400 μg/ml, Sigma, USA) at 34 °C in a saturated humidified incubator. Trypan blue staining was used to determine the Cell viability.
Quantitative real-time PCR
Total RNA was extracted from clinical specimens and OS cell lines and hFOB1.19 cells using RNeasy Plus Mini Kit (Life Technology, US) according to the manufacturer’s instructions. The RNA concentration and purity was determined using an ND-1000 spectrophotometer and stored at −80 °C. Then, cDNA of miRNA was synthesized from 100 ng of total RNA using TaqManH MicroRNA Reverse Transcription (ABI, CA) and cDNA of RNA was synthesized from TaqMan 1 μg of total RNA using Reverse Transcription Reagents (ABI, CA). MiRNAs were reverse-transcribed using specific RT primers; mRNAs were also reverse-transcribed by using random hexamers. The primer sequences used were as follows: FOSL2 forward and reverse, 5′-GAGAGGAACAAGCTGGCTGC-3′ and 5′-GCTTCTCCTTCTCCTTCTGC-3′, respectively; miR-143-3p forward and reverse, 5′-GGGGTGAGATGAAGCACTG-3′ and 5′-CAGTGCGTGTCGTGGAGT-3′, respectively; GAPDH forward and reverse, 5′-ACTTTGGTATCGTGGAAGGACTCAT-3′ and 5′-GTTTTTCTAGACGGCAGGTCAGG-3′, respectively; U6 snRNA forward and reverse, 5′-GCTCGGCAGCACATATACTAAAAT-3′ and 5′-CGCTTCACGAATTTGCGTGTCAT-3′, respectively. Real-time quantitative PCR (qRT-PCR) was performed using SYBR Premix ExTaqTM (Invitrogen, US). GAPDH (for mRNAs) or U6 small nuclear RNA (for miRNAs) was used as an internal control. The relative amount of mRNA or miRNA was calculated according to the comparative threshold cycle value (2−ΔΔCt) method. All operations were repeated at least three times.
Lentivirus infection and oligonucleotide transfection
miR-143-3p mimics (5′-UGAGAUGAAGCACUGUAGCUC-3′), complementary chain (5′-GAGCUACAGUGCUUCAUCUCA-3, miR-143-3p inhibitor (5′-GAGCUACAGUGCUUCAUCUCA-3′) were artificially synthesized from GeneChem Company (Shanghai, China). siRNA for FOSL2 was obtained from GeneChem Company (Shanghai, China). pcDNA3.1 (+)-FOSL2 plasmid was consisted of FOSL2 coding sequences and pcDNA3.1 (+). Transfection of oligonucleotide and plasmids was performed using Lipofectamine 2000 reagent (Invitrogen, USA). The lentivirus vectors which could express miR-143-3p and control vector were obtained from GeneChem Company (Shanghai, China). The lentiviral vector and its packaging vectors were co-transfected into 293T packaging cells to obtain indicated lentivirus. Target cells were infected with fresh lentivirus-containing medium. Stably transfected cells (LV- miR-143-3p) were selected by antibiotic resistance (puromycin, 2 μg/mL) for two weeks.
Luciferase assay
We used the pGL3-reporter luciferase vector to construct the pGL3-FOSL2 3′UTR or pGL3-FOSL2 3′UTR-mut vectors. Vectors expressing miR-143-3p were co-transfected to examine the Luciferase activity. After transfection of 48 h, the Luciferase activity were measured using Dual luciferase reporter assay system (Promega, USA). The ratio of Firefly to Renilla luciferase activity was used as the relative activity of luciferase. All operations were repeated at least three times.
Cell proliferation, colony formation, cell cycle and apoptosis assays
OS cells with different processing methods in different experimental groups were digested and seeded in 96 well plates. Using the CCK-8 assay to detect the cell growth and drawing cell growth curve according to the protocol of the CCK-8 kit (Dojindo, Japan). Cell cycle and apoptosis was analyzed by flow cytometer (BD, USA). For cell cycle analysis, cells were harvested and treated with RNase A (300 μg/ml, Sigma-Aldrich, USA), and then stained with propidium iodide (PI) (10 μg/ml, Sigma-Aldrich, USA). The stained nuclei were measured by flow cytometer. We used Annexin V-FITC/PI kit (BD, USA) to analysis and quantify the cell apoptotic. According to the manufacturer’s protocol. Cells were washed and resuspended at a concentration of ~1 × 106 cells/ml. After treated with Annexin V-FITC and PI solution, cells were stained by Annexin V-FITC/PI for 10~15 min, away from light. Flowjo software was used to determine the apoptosis rate.
In vitro wound-closure assay
OS cell lines suspension (5 × 105/ml) was seeded on 6-well plates and incubated overnight at 37 °C, 5%CO2 incubator. The experiment was divided into 4 groups: miR-143-3p-mi group, miR-143-3p-NC group, miR-143-3p-in group and miR-143-3p-in-NC group. After cell attachment, transfection was performed. All operations were performed without RNA enzymes. 500 μl transfection solution was added to each well. Using a 200 μl pipette tip drew a straight line vertically at the center of each well, and the healing of 0 and 36 h was recorded under the microscope and the healing area of the wound was recorded. Each experiment was repeated three times.
Transwell invasion assays
Cell invasion assays was performed using the Transwell membranes (Corning Inc., New York, NY, USA). Transwell invasion assays were performed according to the manufacturer’s protocol. Briefly, cells were seeded in the upper chamber of the Transwell invasion system and the low chambers were filled with culture medium as chemoattractant and then stay for 48 hours. The cells that invaded into the lower chamber were stained and quantitated by counting under microscopy and taking pictures. The cell number was calculated by Image J software.
Protein extraction and western blotting analysis
Total cell lysates were prepared with a detergent lysis buffer. Western blots were performed as instructions. Anti-FOSL2 and anti-GAPDH (CST, Beverly, MA, USA) were used as first antibody. After washing with PBST buffer, membranes were incubated with secondary antibodies for 2 h. Finally, an enhanced chemiluminescence substrate were used to treat the membranes. GAPDH was used as the loading control.
Mouse models
Four-week-old BALB/c nude mice were obtained from Beijing Weitong Lihua Experimental Animal Technology Co. Ltd. and maintained at Center for Animal Experiment, Renmin Hospital of Wuhan University. The animal experiments were approved by the Institutional Review Board of the Renmin Hospital of Wuhan University and were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. 5 × 106 cells /0.2 ml of 143B-vector or 143B-miR-143-3p cells were injected subcutaneously into the flanks of mice. Each group was randomly assigned to 6 nude mice. The tumor was measured with a caliper once a week for up to 4 weeks. To compare the survival of different mice, 5 × 105 cells /0.2 ml were inoculated subcutaneously and mice survival were also recorded up to 100 days. The tumor volume (V) (mm3) was calculated as the formula: V = (W2 × L)/2. And the curve of tumor growth was depicted 4 weeks after inoculation.
Statistical analysis
SPSS 22.0 software was used for statistical analysis. Independent sample Student’s t-test was used if the quantitative data between groups show normal distribution. If not consistent with the normal distribution, using the Wilcoxon-Mann-Whitney test. The survival curve was calculated using the Kaplan-Meier method and statistical significance was determined by the Log-Rank test. P < 0.05 indicated that the difference was statistically significant.
References
Yu, W. et al. A review and outlook in the treatment of osteosarcoma and other deep tumors with photodynamic therapy: from basic to deep. Oncotarget 8, 39833 (2017).
Bielack, S. S., Hecker-Nolting, S., Blattmann, C. & Kager, L. Advances in the management of osteosarcoma. F1000 Res 5, 2767 (2016).
Friebele, J. C., Peck, J., Pan, X., Abdel-Rasoul, M. & Mayerson, J. L. Osteosarcoma: A Meta-Analysis and Review of the Literature. Am J Orthop (Belle Mead NJ) 44, 547 (2015).
Chang, L., Shrestha, S., LaChaud, G., Scott, M. A. & James, A. W. Review of microRNA in osteosarcoma and chondrosarcoma. Med Oncol 32, 613 (2015).
Taran, S. J., Taran, R. & Malipatil, N. B. Pediatric Osteosarcoma: An Updated Review. Indian J Med Paediatr Oncol 38, 33 (2017).
Evola, F. R. et al. Biomarkers of Osteosarcoma, Chondrosarcoma, and Ewing Sarcoma. Front Pharmacol 8, 150 (2017).
Chen, R., Wang, G., Zheng, Y., Hua, Y. & Cai, Z. Long non-coding RNAs in osteosarcoma. Oncotarget 8, 20462 (2017).
Iorio, M. V. et al. MicroRNA signatures in human ovarian cancer. Cancer Res 67, 8699 (2007).
Li, Z., Yu, X. & Shen, J. Long non-coding RNAs: emerging players in osteosarcoma. Tumour Biol 37, 2811 (2016).
Garajova, I. et al. Non-Coding RNAs as Predictive Biomarkers to Current Treatment in Metastatic Colorectal Cancer. Int J Mol Sci 18 (2017).
David, E. et al. Oncostatin M is a growth factor for Ewing sarcoma. Am J Pathol 181, 1782 (2012).
Wang, K., Jiang, W., Cheng, C., Li, Y. & Tu, M., Pathological and Therapeutic Aspects of Long noncoding RNAs in osteosarcoma. Anticancer Agents Med Chem (2017).
Yang, Y., Wang, S. & Li, T. Altered long non-coding RNAs predict worse outcome in osteosarcoma patients: evidence from a meta-analysis. Oncotarget 8, 35234 (2017).
Shankar, G. M. et al. The role of revision surgery and adjuvant therapy following subtotal resection of osteosarcoma of the spine: a systematic review with meta-analysis. J Neurosurg Spine 27, 97 (2017).
Wang, L., Ge, L. & Yang, X. Combination of subxiphoid and intercostal uniportal video-assisted thoracoscopic surgery for bilateral lung lesions: report of two cases and review of the literature. Postgrad Med 129, 513 (2017).
Sampson, V. B., Yoo, S., Kumar, A., Vetter, N. S. & Kolb, E. A. MicroRNAs and Potential Targets in Osteosarcoma: Review. Front Pediatr 3, 69 (2015).
Cheng, D. et al. MicroRNAs with prognostic significance in osteosarcoma: a systemic review and meta-analysis. Oncotarget (2017).
Chen, Y., Xu, S. F., Xu, M. & Yu, X. C. Intentional marginal resection of periosteal osteosarcoma in combination with neoadjuvant chemotherapy: A report of two cases and a review of the literature. Oncol Lett 13, 1343 (2017).
Anoop, T. M., Geetha, N., Babanrao, S. A. & Jayasree, K. Primary osteosarcoma of rib mimicking lung mass with secondary aneurysmal bone cyst formation. J Thorac Oncol 9, 738 (2014).
Wang, C. et al. Macrophage migration inhibitory factor promotes osteosarcoma growth and lung metastasis through activating the RAS/MAPK pathway. Cancer Lett 403, 271 (2017).
Zandueta, C. et al. Matrix-Gla protein promotes osteosarcoma lung metastasis and associates with poor prognosis. J Pathol 239, 438 (2016).
Akao, Y., Nakagawa, Y., Kitade, Y., Kinoshita, T. & Naoe, T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci 98, 1914 (2007).
Yang, F., Xie, Y. Q., Tang, S. Q., Wu, X. B. & Zhu, H. Y. miR-143 regulates proliferation and apoptosis of colorectal cancer cells and exhibits altered expression in colorectal cancer tissue. Int J Clin Exp Med 8, 15308 (2015).
Cecene, G. et al. Circulating miR-195 as a Therapeutic Biomarker in Turkish Breast Cancer Patients. Asian Pac J Cancer Prev 17, 4241 (2016).
Yang, G. et al. Upregulation of miR-195 increases the sensitivity of breast cancer cells to Adriamycin treatment through inhibition of Raf-1. Oncol Rep 30, 877 (2013).
Ngalame, N. N., Makia, N. L., Waalkes, M. P. & Tokar, E. J. Mitigation of arsenic-induced acquired cancer phenotype in prostate cancer stem cells by miR-143 restoration. Toxicol Appl Pharmacol 312, 11 (2016).
Deftereos, G. et al. Expression of mir-21 and mir-143 in cervical specimens ranging from histologically normal through to invasive cervical cancer. Plos One 6, e28423 (2011).
Wang, L., He, J., Xu, H., Xu, L. & Li, N. MiR-143 targets CTGF and exerts tumor-suppressing functions in epithelial ovarian cancer. Am J Transl Res 8, 2716 (2016).
Ferreira, A. C., Robaina, M. C., Rezende, L. M., Severino, P. & Klumb, C. E. Histone deacetylase inhibitor prevents cell growth in Burkitt’s lymphoma by regulating PI3K/Akt pathways and leads to upregulation of miR-143, miR-145, and miR-101. Ann Hematol 93, 983 (2014).
Kobayashi, M. et al. MicroRNA expression profiling in canine prostate cancer. J Vet Med Sci 79, 719 (2017).
Lui, W. O., Pourmand, N., Patterson, B. K. & Fire, A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 67, 6031 (2007).
Michael, M. Z., O’, C. S., van Holst, P. N., Young, G. P. & James, R. J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res 1, 882 (2003).
Hirahata, M. et al. PAI-1, a target gene of miR-143, regulates invasion and metastasis by upregulating MMP-13 expression of human osteosarcoma. Cancer Med 5, 892 (2016).
Akao, Y. et al. Role of anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer Gene Ther 17, 398 (2010).
Xu, B. et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem 350, 207 (2011).
Kent, O. A., Fox-Talbot, K. & Halushka, M. K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 32, 2576 (2013).
Wang, J. et al. FOSL2 positively regulates TGF-beta1 signalling in non-small cell lung cancer. Plos One 9, e112150 (2014).
Davies, J. S., Klein, D. C. & Carter, D. A. Selective genomic targeting by FRA-2/FOSL2 transcription factor: regulation of the Rgs4 gene is mediated by a variant activator protein 1 (AP-1) promoter sequence/CREB-binding protein (CBP) mechanism. J Biol Chem 286, 15227 (2011).
He, J. et al. miR-597 inhibits breast cancer cell proliferation, migration and invasion through FOSL2. Oncol Rep 37, 2672 (2017).
Wrann, C. D. et al. FOSL2 promotes leptin gene expression in human and mouse adipocytes. J Clin Invest 122, 1010 (2012).
Acknowledgements
This study was supported in part by Grants from the Natural Science Foundation of China (No. 81341078 and 81502575), and Fundamental Research Funds for the Central Universities (2042015kf0069).
Author information
Authors and Affiliations
Contributions
X.S., G.D. and L.Y. carried out all the experiments. X.S. and G.D. wrote the manuscript. L.Y., Q.H. and J.C. participated in the statistical analysis and help to draft the manuscript. X.S., L.Y. and W.G. were responsible for the experimental design. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, X., Dai, G., Yu, L. et al. miR-143-3p inhibits the proliferation, migration and invasion in osteosarcoma by targeting FOSL2. Sci Rep 8, 606 (2018). https://doi.org/10.1038/s41598-017-18739-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-18739-3
This article is cited by
-
Hsa-miR-143-3p inhibits Wnt-β-catenin and MAPK signaling in human corneal epithelial stem cells
Scientific Reports (2022)
-
Distinct power of bone marrow microRNA signatures and tumor suppressor genes for early detection of acute leukemia
Clinical and Translational Oncology (2022)
-
CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression
Journal of Experimental & Clinical Cancer Research (2021)
-
Circular RNA circCSNK1G3 induces HOXA10 signaling and promotes the growth and metastasis of lung adenocarcinoma cells through hsa-miR-143-3p sponging
Cellular Oncology (2021)
-
microRNA-143-3p contributes to inflammatory reactions by targeting FOSL2 in PBMCs from patients with autoimmune diabetes mellitus
Acta Diabetologica (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.