Abstract
Connective tissues (CT) support and connect organs together. Understanding the formation of CT is important, as CT deregulation leads to fibrosis. The identification of CT specific markers has contributed to a better understanding of CT function during development. In developing limbs, Osr1 transcription factor is involved in the differentiation of irregular CT while the transcription factor Scx labels tendon. In this study, we show that the CXCL12 and CXCL14 chemokines display distinct expression pattern in limb CT during chick development. CXCL12 positively regulates the expression of OSR1 and COL3A1, a collagen subtype of irregular CT, while CXCL14 activates the expression of the tendon marker SCX. We provide evidence that the CXCL12 effect on irregular CT involves CXCR4 receptor and vessels. In addition, the expression of CXCL12, CXCL14 and OSR genes is suppressed by the anti-fibrotic BMP signal. Finally, mechanical forces, known to be involved in adult fibrosis, control the expression of chemokines, CT-associated transcription factors and collagens during limb development. Such unexpected roles of CXCL12 and CXCL14 chemokines during CT differentiation can contribute to a better understanding of the fibrosis mechanisms in adult pathological conditions.
Similar content being viewed by others
Introduction
In the body, the main role of connective tissues (CT) is to support organs and to connect cells and tissues. CT are primarily composed of extracellular matrix produced by fibroblasts that derive from mesenchymal progenitor cells during development. Several types of CT exist: specialized CT refers to bone and cartilage, while dense CT is divided into regular CT, which corresponds to tendon and ligament and irregular CT (ICT). ICT includes CT surrounding organs, as for example the perichondrium around cartilage and the epimysium around muscles, and CT inside organs, as for example muscle connective tissue (MCT)1. Understanding the specification and differentiation processes of CT types from undifferentiated mesenchymal cells is important, as CT deregulation leads to fibrosis, a process attributed to excess deposition of extracellular matrix in response to injury, inflammation or aging2. Fibrosis is also a major pathological feature of chronic autoimmune diseases, tumour invasion and progressive myopathies2.
Until a few years ago, studying the specification of CT types during embryogenesis was still challenging, due to the absence of specific markers to distinguish the different forms of CT. Most studies designed to identify CT-specific genes during development have been conducted on the vertebrate limb musculoskeletal system as a model. Indeed, this multicomponent structure formed by skeletal muscle, bone and cartilage (specialized CT), tendon and ligament (regular CT) and irregular CT (ICT), surrounding and connecting the different elements of the musculoskeletal system, is particularly suitable to investigate CT differentiation in the embryo. In limbs, all types of CT derive from the lateral plate mesoderm3,4, while myogenic cells originate from the somitic dermomyotome and migrate into the limb buds3,5. Classical embryological approaches have shown that limb lateral plate mesoderm contains positional information cues for limb formation6,7. The limb mesenchyme also influences muscle patterning8, highlighting the importance of CT derivatives for limb musculoskeletal morphogenesis. In the developing limb, studies on CT differentiation first focused on collagen, the major component of the extracellular matrix and showed that type I and type III collagens are both expressed in dense regular and irregular CT9. However, type I collagen becomes progressively predominant in tendons, while both type III and type VI collagens mostly characterize both ICT and MCT9,10,11.
More recently, the identification of specific markers and genetic tools allowing the labelling and manipulation of CT fibroblasts has largely contributed to a better understanding of the role of CT types during limb development. The Scleraxis (Scx) gene, encoding a bHLH transcription factor, is expressed in tendon and ligament cells12 and Scx−/− mice showed severe defects in force-transmitting tendons13. Scx consequently characterizes dense regular CT (tendon and ligament). In addition, Scx has been shown to activate Col1a1 transcription, coding for one chain of the main collagen expressed in tendon cells14. In contrast to tendon and ligament, the characterization of ICT and MCT remains challenging. Tcf4, a member of the Tcf/Lef family of transcription factors, is highly expressed in MCT during chick and mouse limb development15,16. TCF4 gain- and loss-of-function experiments demonstrated that TCF4-expressing cells contribute to limb muscle patterning in chick embryos15. Similarly, mispatterning of limb muscles and tendons are observed after ablation of the T-box transcription factor Tbx5 in mouse lateral plate-derived cells17. Interestingly, Scx and Tcf4 genes have been shown to be critical regulators of fibrosis in different adult organs18,19. However, although Tcf4 and Tbx5 are considered as MCT-associated markers, Tcf4 is also expressed in myogenic cells16 and Tbx5 is also observed in cartilage, tendon and muscle progenitors of mouse limbs20, showing that both factors are not specific to ICT and MCT. More recently, the zinc finger transcription factors Odd Skipped-related-1 and -2 (Osr1 and Osr2) have been identified as being expressed in ICT during limb development, with a prevalence of Osr2 expression in MCT21. Osr1 and Osr2 genes are not expressed in tendon, ligament and cartilage, with the exception of the presumptive joints21. Both Osr1 and Osr2 drive ICT differentiation at the expense of cartilage and tendon and are required for the differentiation of ICT fibroblasts22. Overexpression of OSR1 or OSR2 in chick limb mesenchymal cells induces the expression of ICT markers such as COL3A1 and COL6A1, while downregulating the expression of cartilage and tendon markers. Conversely, OSR1 or OSR2 inactivation downregulates COL3A1 and COL6A1 expression, while increasing cartilage formation in chick limb cells22.
During limb development, the expression of these CT-associated transcription factors is regulated by extrinsic signalling pathways. BMP4 represses Tcf4 23 and Scx expression12, while SCX expression is positively regulated by FGF424,25. Importantly, both BMP and FGF signalling pathways are involved in the regulation of adult tissue fibrosis26,27. Chemokines are also important regulators of fibrosis28 and interestingly, CXCL12 and CXCL14 have been shown to delineate some CT subpopulations in embryonic limbs29,30,31. Identified CXCL12 functions during embryogenesis include essential roles in the migration process of hematopoietic stem cells32,33, neurons34,35, germ cells36,37 and skeletal muscle cells29. These functions are mediated by two chemokine receptors, CXCR4 and CXCR7 that signal individually in different cells or act in a cooperative manner in the same cell38. The role of CXCL14 during development remains more elusive but CXCL14 possesses chemoattractive activity for activated macrophages, immature dendritic cells and natural killer cells39 and it has been shown that CXCL14-forced expression suppresses tumour growth in mice40,41. Specific receptors for CXCL14 have not yet been identified but it is proposed that CXCL14 is a natural inhibitor of the CXCL12/CXCR4 axis42.
In this study, we show that CXCL12 and CXCL14 chemokines display distinct expression pattern in limb CT and regulate the expression of specific CT markers. We provide evidence that CXCL12 positively regulates the expression of OSR1 and COL3A1, a major collagen subtype of the ICT, in chick embryonic limbs and fibroblasts, while CXCL14 activates the expression of the tendon marker SCX in chick fibroblasts. Moreover, the expression of CXCL12, CXCL14 and OSR genes is negatively regulated by BPM4, while the blockade of BMP activity is sufficient for enhancing CXCL12, CXCL14 and OSR1 expression. Lastly, the expression of CXCL12, CXCL14 and CT markers is decreased in the absence of muscle contraction.
Results
CXCL12 and CXCL14 chemokines display distinct expression pattern in limb CT during chick development
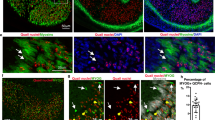
We analysed the expression patterns of CXCL12 and CXCL14 in chick limbs at different developmental stages. Comparison was made with OSR1, OSR2 and PDGFR-α, three ICT-associated markers and with SCX for tendon CT. Limb differentiated muscles were visualized by myosin expression (MF20 antibody). At E5 of development, CXCL12 displayed a diffuse expression in limb CT, with a strong expression in ICT between cartilage elements, overlapping with OSR2 expression (Fig. 1A,C). CXCL12 was also expressed in MCT (Fig. 1D), overlapping with OSR1 and OSR2 expression (Fig. 1D–F). CXCL12 also colocalised with PDGFR-α, which appeared widely expressed in the forelimb ICT and at a lower level in MCT (Fig. 1G–I), as previously described43,44. At E5, CXCL14 was expressed in forelimb ectoderm and faintly in proximal CT (Fig. 1J, arrow), in a region where CXCL12 was also expressed (Fig. 1K, arrow). CXCL14 exhibited a partial overlap with OSR1 and SCX expression but not with that of OSR2 in proximal limb regions (Fig. 1J,L–N, arrows). From E6, in addition to ectoderm expression, CXCL14 was expressed in a subpopulation of ICT mostly located in limb ventral regions (Fig. 1O–Q). At E7, CXCL12 expression was still observed in ICT and MCT, overlapping with OSR1 expression in ICT and both OSR1 and OSR2 expression in MCT (Fig. 2A–F). At this stage, CXCL12 expression partially overlapped with COL1A1 but not with SCX (Fig. 2G–I). At E10, CXCL12 expression was reminiscent of that of PDGFR-α in ICT and MCT (Fig. 2J–L). In addition to ectodermal expression, CXCL14 was observed in ICT surrounding cartilage (Fig. 2M), but also in MCT of a subgroup of ventral muscles (Fig. 2M,P) and in ICT at the vicinity of SCX expression (Fig. 2R,S). At this stage, CXCL14 and CXCL12 expression did not obviously overlapped in ICT, but partially colocalised around cartilage and in MCT (Fig. 2M,N).
Chemokine expression in relation to CT markers in forelimb of E5 and E6 chick embryos. In situ hybridizations for CXCL12 (A,D,K), CXCL14 (J,O,P,Q), OSR1 (B,E,L), OSR2 (C,F,M) and immunohistochemical detections for PDGFRα (G,I), MF20 (A–F,H,I,P,Q) on serial transverse (A–I,Q) or longitudinal (J–P) sections. CXCL12 is expressed in CT and MCT partially overlapping with OSR1, OSR2 and PDGFRα expression. CXCL14 exhibits a restricted expression in CT (J,O–Q), partially overlapping with CXCL12, OSR1 and SCX expression (arrows in J,K,L,N). (D,E,F,I) represent high magnifications of the squared regions respectively in (A,B,C and G,H,D): dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. Bars: 200 µm in (A–C,G,H,J–O,Q); 100 µm in (D–F,P).
Chemokine expression in relation to CT markers in forelimb of chick embryos from E7 to E10. In situ hybridizations for CXCL12 (A,D,G,J,N), CXCL14 (M,P,R,Q), OSR1 (B,E), OSR2 (C,F), COL1A1 (H), SCX (I,O,S) and immunohistochemical detections for PDGFRα (K,L), MF20 (A–F, J–L,P–S) on serial transverse (A–L, P,Q) or longitudinal (M–O,R,S) sections. CXCL12 is expressed in CT and MCT partially overlapping with OSR1, OSR2 and PDGFRα expression but with no evident colocalisation with COL1A1 or SCX expression. CXCL14 expression partially overlaps with CXCL12 expression in CT and MCT and is observed in the vicinity of SCX expression in CT giving rise to tendon. (D,E,F,L,R,S) represent high magnifications of the squared regions respectively in (A,B,C,K,N,O). D: dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. Bars: 200 µm in (A–C,G–I,M–P); 100 µm in (D–F,J,K); 50 µm in (Q–S).
Taken together, our results show that CXCL12 and CXCL14 chemokines exhibit distinct and dynamic expression patterns during chick limb development in ICT and MCT. CXCL12 expression is widespread in limb ICT and MCT, while CXCL14 is expressed in the vicinity of a subset of tendons and restricted to MCT of specific muscles, mostly located ventrally in limbs. The comparison of CXCL12 and CXCL14 expression with that of limb CT-markers shows a good correlation between the expression of CXCL12 and OSR transcription factors, while CXCL14 expression appears complementary in some limb regions to the tendon CT-marker SCX, suggesting a differential role for the two chemokines in limb CT regulation.
CXCL12 and CXCL14 regulate the expression of different CT markers during chick limb development
To test the involvement of CXCL12 and CXCL14 in limb CT differentiation, we performed CXCL12 and CXCL14 gain-of-function experiments in vitro and in vivo. We first overexpressed CXCL12 and CXCL14 chemokines in primary cultures of E10 chick embryonic fibroblasts using the replication-competent RCAS retrovirus system. Chick embryonic fibroblasts were transfected with recombinant retroviruses that spread in dividing cells, allowing a general expression of the gene of interest after 4 days of culture (Fig. 3A). CXCL12 overexpression in chick embryonic fibroblasts led to a significant increase in the mRNA levels of OSR1, OSR2 and COL3A1 genes, while the expression of COL1A2, COL6A1, PDGFRA and SCX was not significantly modified (Fig. 3B). CXCL14 overexpression significantly increased the mRNA levels of the tendon marker SCX, but did not change the expression levels of the other CT-associated markers (Fig. 3C). We next overexpressed CXCL12 and CXCL14 in ovo, by grafting RCAS-CXCL12 or RCAS-CXCL14 producing fibroblasts into limb buds of E4 chick embryos to allow ectopic gene expression in limb regions24 (Fig. 4A). Embryos were collected at E10 and the expression of CT markers was analysed by in situ hybridization to transverse limb sections (Fig. 4A). Ectopic CXCL12 expression (Fig. 4Bb,h) resulted in an increase of OSR1 and COL3A1 expression in the infected limb regions (Fig. 4Bd,f,j,l) compared to control limbs (Fig. 4B). We did not observe any obvious change of OSR2 expression by in situ hybridization in the RCAS-CXCL12-infected regions (data not shown). CXCL14 overexpression did not induce ectopic SCX expression in chick forelimbs when compared to control limbs (Fig. 4C), although we cannot exclude an increase of SCX expression in tendons.
Overexpression of CXCL12 and CXCL14 chemokines increases the expression of CT markers in vitro. (A) Experimental scheme of in vitro transfection of chicken embryonic fibroblasts (CEF) by RCAS viruses expressing chick CXCL12 or CXCL14 constructs. (B,C) RT-qPCR analyses of the expression level of CT markers in CEF overexpressing CXCL12 (B, n = 6) or CXCL14 (C, n = 6) showing that CXCL12 upregulates significantly OSR1, OSR2 and COL3A1, while CXCL14 increases SCX. For each gene, the mRNA levels of control cultures (n = 6) were normalised to 1. P values were analysed by two-tail and unpaired Student’s t-test using Microsoft Excel. **P < 0.01; ***P < 0.001; Error bars indicate s.d.
Overexpression of CXCL12 and CXCL14 chemokines in vivo increases the expression of CT markers in the chick embryonic forelimb. (B) Ectopic expression of CXCL12 upregulates OSR1 and COL3A1 expression (n = 6). In situ hybridizations for CXCL12 (a,b,g,h), OSR1 (c–f) and COL3A1 (i–l) on serial transverse sections of control (a,c,e,g,i,k) and grafted (b,d,f,h,j,l) E10 chick forelimbs. (e,f,k,l) represent high magnifications of the squared regions respectively in (c,d,i,j). (C): Ectopic expression of CXCL14 does not modify SCX expression (n = 6). In situ hybridizations for CXCL14 (a,b) and SCX (c,d) on serial transverse sections of control (a,c) and grafted (b,d) E10 chick forelimbs. (D) dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. Bars: 200 µm in B (a-d, g–j), C (a–d); 100 µm in B (e,f,k,l).
Altogether, these results show that CXCL12 and CXCL14 chemokines differentially regulate CT-associated markers in vivo and in vitro. CXCL12 positively regulates the expression of the ICT markers OSR1 and COL3A, while CXCL14 regulates the tendon marker SCX, at least in vitro.
Misregulation of CXCL12/CXCR4 axis in limb alters vascular network
In order to investigate which receptor could be involved in the effect of CXCL12 on ICT gene expression, we first analysed the expression of the two recognized CXCL12 receptors, CXCR4 and CXCR738, in developing chick limbs. At E5, CXCR4 was expressed in endothelial cells labelled with MEP21 (Supp. Fig. 1A–C), as already described in chick embryos45,46, while CXCR7 was observed in limb ICT, with a strong expression in ICT surrounding cartilage elements (Supp. Figure 1A–C). At E10, CXCR7 appeared expressed in some regions of ICT around muscles, while CXCR4 was still expressed in endothelial cells at this stage (Supp. Fig. 1D–F).
Consistent with the endogenous CXCR4 expression in endothelial cells of developing chick limbs (Supp. Fig. 1) and adults47, the CXCL12/CXCR4 axis is known to be involved in vasculogenesis during development and in pathological conditions48,49. We thus analysed the consequences of CXCL12 overexpression for limb vascular network in chick embryos. CXCL12 was overexpressed in limb CT by electroporating a PT2AL-CMV-TOMATO-T2A-CXCL12 construct in chick limb lateral plate mesoderm (Fig. 5A) using the Peptide 2 A system allowing simultaneous expression of Tomato and the gene-of -interest50. Limb lateral plate mesoderm was electroporated at E2.5 and embryos were collected at E8 for ink injection in the heart. As expected, CXCL12 overexpression in limb CT visualized with Tomato fluorescence affected the vascular network in electroporated wings (Fig. 5Bb,c), when compared to controls (Fig. 5Ba). The vascular phenotype was characterized by a more or less enlargement of limb vessels and abnormal vascular branching upon CXCL12 overexpression (Fig. 5Bb,c). We next investigated the distribution of endothelial cells after CXCL12 overexpression, by analysing CXCR4 and MEP21 expression. Retroviral overexpression of CXCL12 (Fig. 5C) modified the distribution of CXCR4- and MEP21-positive cells in the infected limb regions (Fig. 5Dd–f,i,j) compared to control limbs (Fig. 5Da–c,g,h), confirming that increased CXCL12 expression in ICT resulted in an abnormal vascularisation in chick limbs.
Overexpression of CXCL12 affects limb vessel network and distribution of CXCR4-positive endothelial cells. (A) Experimental scheme of the electroporation procedure of PT2AL-CMV-TOMATO-T2A-CXCL12 construct in the chick limb lateral plate mesoderm. (B) Ectopic expression of CXCL12 in the lateral plate mesoderm resulted in abnormal limb vascular network in electroporated wings (b,c, n = 5/8) when compared to control wings (a, n = 8). (a,b) E8 wings after black ink injection. (c) Tomato expression in corresponding electroporated wing. (C) Experimental scheme of the grafting procedure of CEF pellets expressing chick CXCL12 constructs in chick embryonic forelimbs. (D) Ectopic expression of CXCL12 modifies the distribution of CXCR4- and MEP21-positive cells in chick limbs (n = 6). In situ hybridization for CXCL12 (a,d,g,i) and immunochemistry for CXCR4 (b,c,e,f) and MEP21 (h,j) on serial transverse sections of control (a–c,g,h) and grafted (d–f,I,j) E8 chick forelimbs. (c,f) represent high magnifications of the squared regions respectively in (b,e). D dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. Bars: 1 mm in B (a–c), 200 µm in D (a,b,d,e), 100 µm in D (c,f) 50 µm in D (g–j).
To test the possible involvement of CXCR7 in the CXCL12 effect on CT-associated genes, we overexpressed a dominant-negative form of CXCR7 (dn-CXCR7) in ovo, by grafting RCAS-dn-CXCR7 producing fibroblasts into limb buds of E4 chick embryos. Embryos were collected at E10 to analyse the expression of CT markers by in situ hybridization to transverse limb sections. Ectopic dn-CXCR7 expression (Supp. Fig. 2B,E,H,K) did not modify the expression of the transcription factors OSR1, OSR2 and SCX nor that of COL3A1 gene in infected limb regions (Supp. Fig. 2C,F,I,L) compared to controls (Supp. Fig. 2A,G,J). This indicates that CXCR7 receptor does not mediate the CXCL12 effect on the expression of CT markers.
Taken together, these data suggest that CXCL12 effect on ICT gene expression involves CXCR4 but not CXCR7 receptors.
CXCL12 and CXCL14 expression is not regulated by FGF4
During development, FGF4 positively regulates SCX expression24,51 and the expression of ICT collagens, including COL1A1, COL3A1 and COL6A1 25 in chick limbs. Interestingly, FGF has been shown to be involved in fibrosis regulation in different pathological situations27. We then analysed the effects of FGF4 overexpression on CXCL12 and CXCL14 expression in chick limbs. FGF4 beads were implanted into limbs at E4.5 and fixed 48 hours later (Supp. Figure 3A). As previously shown24,25,51, SCX expression was strongly induced at the vicinity of FGF4 beads in grafted limbs compared to normal SCX expression in control limbs (Supp. Fig. 3Ba,b,g,h). However, no ectopic expression of CXCL12 and CXCL14 was observed in ectopic SCX expression domains (Supp. Fig. 3Ba–f). In addition, we did not observe any OSR1 and OSR2 activation around FGF4 beads (Supp. Fig. 3Bg–l). However, an inhibition of OSR1 and OSR2 expression was observed in the vicinity of FGF4 beads (Supp.Fig. 3Bh,j,l) compared to control limbs (Supp. Fig. 5Bg,i,k). These results show that ectopic expression of FGF4 does not modify CXCL12 and CXCL14 expression but leads to the inactivation of both OSR1 and OSR2 genes in chick limbs, while increasing SCX expression. It is likely that OSR gene downregulation observed after FGF4 bead grafts is independent of CXCL12 and CXCL14 chemokines. However, since OSR1 and OSR2 overexpression inhibits SCX expression in chick limb cells22, these results suggest that SCX and OSR genes mutually repress each other in mesenchymal cells during chick limb development.
The anti-fibrotic BMP signalling pathway negatively regulates the expression of CXCL12 and CXCL14
BMP signalling has been demonstrated as an antagonist of fibrosis in various organs52 but very few studies have investigated its potential role in the regulation of CT types during development. Conversely to FGF4, BMP4 inhibits SCX expression in chick embryonic limbs12,53. We then looked at the possible effect of BMP signalling on CXCL12 and CXCL14 expression in vivo by overexpressing BMP4 in chick limbs. Pellets of transfected RCAS-mBmp4 fibroblasts were grafted into E4 limbs, which were collected at E8 to analyse the expression of chemokines and CT markers (Fig. 6A). Bmp4 overexpression resulted in a downregulation of CXCL12 and CXCL14 expression (Fig. 6Bd–f) and a concomitant decrease in both OSR1 and OSR2 expression in limb mesenchyme (Fig. 6Bj–l), compared to control (Fig. 6Ba–c,g–i). These results were supported in vitro by the analysis of mRNA levels of chemokines and CT markers in embryonic fibroblasts overexpressing Bmp4, which showed downregulation of CXCL12, CXCL14, OSR1 and OSR2 expression (Fig. 6C). These data evidence that BMP4 gain-of-function experiments lead to a decrease of chemokine and CT marker expression in chick limb cells. In order to test if BMP loss-of-function experiments would provoke the opposite result, BMP signalling pathway was inhibited in vivo by overexpressing the BMP antagonist Noggin in chick limbs54. Pellets of transfected RCAS-cNOGGIN producing fibroblasts were grafted into E4 limbs, which were collected at E9 to analyse the expression of chemokines and CT markers (Fig. 7A). NOGGIN overexpression led to an upregulation of CXCL12 and CXCL14 expression in both ICT and MCT (Fig. 7Ba,b,f,g) and a concomitant increase in OSR1 expression (Fig. 7Bh), compared to control limbs (Fig. 7Bc–e). In this experimental design, OSR2 and SCX expression was not obviously modified (data not shown).
BMP signaling pathway down-regulates the expression of chick CT markers in vivo and in vitro. BMP signalling represses CXCL12, CXCL14, OSR1, and OSR2 expression in the chick forelimb CT and MCT (n = 5). (A) Experimental scheme of the grafting procedure of CEF pellets expressing mouse Bmp4 construct in the chick embryonic forelimb. (B) In situ hybridizations for CXCL12 (b,e), CXCL14 (c,f), OSR1 (h,k), OSR2 (i,l), mBmp4 (d,j) and immunohistochemical detection for MF20 (a,d,g,j) on serial transverse sections of control (a–c,g–i) and grafted (d–f,j–l) E8 chick forelimbs. The squared regions in (a,d,g,j) delineate the high-magnified parts of the adjacent sections revealed for chemokines or CT markers. D: dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. Bars: 200 µm in B (a,d,g,j), 100 µm in B (b,c,e,f,h,k,i,l). (C): RT-qPCR analyses of expression level of readout targets of BMP4 signalling and of CT markers in CEF expressing RCAS-mBmp4 construct, showing that BMP4 expression significantly decreases CXCL12, CXCL14, OSR1 and OSR2 expression (n = 6). For each gene, the mRNA levels of control cultures (n = 6) were normalised to 1. P values were analysed by two-tail and unpaired Student’s t-test using Microsoft Excel. ***P < 0.001; Error bars indicate s.d.
Inhibition of BMP signalling activates the expression of chick CT markers in vivo. BMP inhibition increases CXCL12, CXCL14 and OSR1 expression in the chick forelimb CT and MCT (n = 4). (A) Experimental scheme of the grafting procedure of CEF pellets expressing chick NOGGIN construct in the chick embryonic forelimb. (B) In situ hybridization for NOGGIN (a,b), CXCL12 (c,f), CXCL14 (d,g), OSR1 (e,h) and immunohistochemical detection for MF20 (c,f) on serial transverse sections of control (c–e) and grafted (f–h) E9 chick forelimbs. The squared regions in a delineate the high-magnified region of the adjacent sections revealed for NOGGIN, chemokines or CT markers. Arrows indicate the expression in CT and MCT. Arrowheads indicate the expression in perichondrium. D: dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. Bars: 200 µm in A (b–h), 100 µm in A (a).
Taken together, these data show that the anti-fibrotic factor BMP4 negatively regulates the expression of CXCL12 and CXCL14 chemokines and OSR1 and OSR2 transcription factors, while BMP inhibition is sufficient to induce the expression of CXCL12, CXCL14 and OSR1 in chick limbs.
Mechanical forces act upstream CXCL12 and CXCL14 to regulate CT markers
The musculoskeletal system is sensitive to mechanical loads exerted by muscle contractions. Mechanical forces generated by muscle activity have been shown to be crucial for the formation of components of the musculoskeletal system during development51,55,56. In the adult, studies indicate that mechanical forces contribute to fibrosis by regulating CT fibroblasts in bone, cartilage and interstitial tissues of most organs57. During development, SCX expression is sensitive to mechanical forces in tendons of mouse and chick limbs51,58 but no investigation has been conducted concerning the role of mechanical signals on ICT and MCT during development. To assess the effect of mechanical forces in this context, chick embryos were immobilised using the decamethonium-bromide (DMB) pharmacological agent, an acetylcholine agonist leading to rigid muscle paralysis and immobilisation of the embryo59. DMB or control solutions were applied in E4.5 embryos and limbs were collected 2 or 3 days later to analyse the expression of chemokines and CT marker by in situ hybridization and RT-qPCR. DMB has been shown to bind motor endplates and block muscle contraction60. As neuromuscular contractions in chick embryonic limbs are effective from E5.561, limb muscles were not grossly affected at E6.5 after 2 days of immobilisation (Fig. 8Ah, l), while paralysis leads to muscle degeneration later during development, as previously described62. CXCL12 and CXCL14 expression was reduced in MCT and ICT surrounding muscles in paralysed limbs (Fig. 8Ab,d). However, CXCL12 expression was maintained in CT surrounding bones, while CXCL14 was still expressed in the ectoderm in immobilised conditions (Fig. 8Ab,d). The expression of OSR1 and OSR2 was also decreased in the absence of mechanical activity compared to control limbs (Fig. 8Ae–j). RT-qPCR analyses confirmed the downregulation of the mRNA levels of CXCL12, CXCL14, OSR1 and OSR2 in paralysed limbs (Fig. 8B) and also showed that the mRNA levels of COL1A2, COL3A1 and COL6A1 genes were significantly reduced in immobilised conditions. The decrease of CT marker expression cannot be attributed to a loss of CT cells since TCF4-positive cells were still observed in limbs of immobilised as compared to control embryos (Fig. 8Ah,l). Taken together, these results show that the expression of CXCL12, CXCL14 and CT markers is sensitive to mechanical forces during limb development.
Mechanical forces act upstream CXCL12 and CXCL14 chemokines to regulate the expression of CT markers in the chick embryonic forelimb. (A) Immobilisation of the chick embryonic forelimb downregulates the expression of CXCL12 and CXCL14 and of the CT markers (n = 3). In situ hybridizations for CXCL12 (a,b), CXCL14 (c,d), OSR1 (e,i), OSR2 (f,j) and immunodetection of MF20 (g,k) and TCF4 (h,l) on serial transverse sections of control (a,c,e–h) and DMB-treated (b,d,i–l) E6.5 chick forelimbs. (D) dorsal, V: ventral, a: anterior, p: posterior, r: radius, u: ulna. (B) RT-qPCR analyses of CXCL12, CXCL14 and CT markers in E7.5 immobilized chick forelimbs (n = 4) showing that both chemokine and CT marker expressions are reduced in immobilisation conditions. For each gene, the mRNA levels of control limbs (n = 4) were normalized to 1. P values were analyzed by two-tail and unpaired Student’s t-test using Microsoft Excel. **P < 0.01; ***P < 0.001; Error bars indicate s.d.
Discussion
Fibrosis is a pathological reaction of CT arising from chronic inflammation, which progressively destroys tissue architecture. It results from the activation of fibroblasts, which produces excessive deposition of extracellular matrix. Chemokines appear to be central mediators of fibrosis initiation and progression63. During development, embryonic fibroblasts are constantly activated to participate to tissue remodelling64. Therefore, understanding fibroblast features and regulation during development is an important issue to address fibrosis mechanisms. In this study, we show that CXCL12 and CXCL14 chemokines positively regulate the expression of different CT-associated markers in chick limb fibroblasts. In addition, we show that BMP signalling, known for its anti-fibrotic role, negatively regulates CXCL12 and CXCL14 expression in limb CT during development. Finally, mechanical forces, which are essential for the progression of fibrosis65, also regulate the expression of CXCL12 and CXCL14 and CT markers.
We show here that the chemokine CXCL12 is sufficient to activate the expression of ICT markers, OSR1 and COL3A1 in chick limb fibroblasts in vitro and in vivo, identifying CXCL12 as a promoting factor of ICT differentiation during chick limb development. We show that, although expressed in some regions of the limb ICT, CXCR7 receptor is not involved in the CXCL12 effect on the expression of CT markers. CXCL12 overexpression in chick limb ICT modifies the distribution of limb endothelial cells expressing CXCR4 receptor and leads to abnormalities in limb vasculogenesis, mostly characterized by the enlargement of some vessels and aberrant vascular branching. Interestingly, CXCL12 has been shown to promote PDGF-B production via direct activation of transcription of Pdgfb gene in endothelial cells66. PDGF-B is known to play a role in CT remodelling and fibrosis67,68 and to induce collagen transcription via PDGFR-α expressed in CT in chick limbs44. Consequently, it can be hypothesized that increasing CXCL12/CXCR4 signalling pathway in chick embryonic limbs modifies the limb vascular network and the secretion of PDGF-B which, in turn, would promote the expression of ICT markers (Supp. Fig. 4). The promoting effect of CXCL12 on ICT during development is consistent with the recognized role of CXCL12/CXCR4 axis in adult fibrosis. CXCL12 expression is increased in different human fibrotic pathological situations, such as idiopathic pulmonary fibrosis69 and chronic pancreatitis70. In human lung and prostate fibroblasts, CXCL12 has been shown to induce, via the CXCR4 receptor, the expression of α-SMA, the key marker of activated fibroblast71,72.
As CXCL12, CXCL14 has been demonstrated as being involved in tissue fibrosis. CXCL14 expression is upregulated in idiopathic pulmonary fibrosis73 and chronic inflammatory arthritis74 and has been shown to stimulate prostate fibroblast proliferation and migration75. Our results show that CXCL14 specifically activates the tendon-CT marker SCX in chick fibroblasts, while not affecting the other CT markers. It has been shown recently that SCX controls fibroblast activation in the heart and could be a potent regulator in fibrotic diseases in the cardiovascular system18. In addition, SCX is activated by the pro-fibrotic factor TGF-β during chick or mouse limb development51,76. Previous experiments have shown that limb ectoderm induces SCX expression in limb mesenchyme at E3.5 but is not required for SCX maintenance after E4.5 of development12. As CXCL14 is expressed in limb ectoderm (Fig. 2,30), one attractive hypothesis would be that the ectodermal-derived CXCL14 regulates SCX expression in limb mesenchymal cells in early limb buds.
We show here that forced-expression of BMP4 decreases CXCL12, CXCL14, OSR1 and OSR2 expression in embryonic fibroblasts and limbs, while the inhibition of BMP signalling by NOGGIN overexpression leads to increased expression of CXCL12, CXCL14 and OSR1 in chick limbs. The anti-fibrotic effects of BMPs has been demonstrated in many organs, where it has been shown that BMP acts by inhibiting the pro-fibrotic activity of TGFβ or directly repressing the expression of Ctgf gene, encoding a pro-fibrotic factor, and Acta2 gene, encoding α-SMA52. Because CXCL12 positively regulates OSR1 expression (Fig. 4), it is tempting to speculate that the modification of OSR1 expression by BMP signalling is a consequence of CXCL12 down- and upregulation following BMP gain- and loss-of-function experiments, respectively. This would indicate a pivotal role for CXCL12 and CXCL14 chemokines as downstream targets of BMP4 signalling pathway to regulate ICT markers during chick limb development (Supp. Figure 4). In bone marrow stromal cell lines, SMAD-binding elements have been identified in the Cxcl12 promoter and BMP4 has been shown to regulate Cxcl12 expression, as treatment of stromal cells with the BMP antagonist Noggin significantly increased Cxcl12 levels77. We show here that BMP overexpression or inhibition decreases or increases, respectively, CXCL14 expression. As SCX expression in chick limbs has been shown to be restricted by localized BMP signalling12, it can be suggested that this restriction could occur through the inhibition of CXCL14 by BMP signalling.
The expression of CXCL12 and CXCL14 chemokines and their CT target genes is decreased in immobilisation conditions. Numerous studies have underlined the role of the mechanical activity as a profibrotic stimulus65. TGF-β, one important factor in fibrosis, is activated as the direct result of mechanical tension induced by the extracellular matrix78. Generation of mechanical tension results in the expression of α-SMA and the increase in matrix deposition in multiple tissues65. Mechanical activity has been also shown to modulate or synergize with Scx to promote the differentiation of mesenchymal cells towards tendon CT79,80. During chick embryonic development, muscleless or aneural limbs deprived of musculoskeletal activity exhibit reduced SCX expression24 and embryo immobilisation results in a FGF4-dependent downregulation of SCX in limbs51. The downregulation of CXCL12 and CXCL14 expression in immobilisation conditions is specific to MCT and ICT surrounding muscles, while not affecting that of cartilage regions (CXCL12) and ectoderm (CXCL14). Among the mechanisms involved in the response of adult muscle to exercise is the activation of the fibrotic TGF-β signalling pathway, which leads to an increase in the expression of CTGF and collagens in MCT81. TGF-β has been shown to control dense regular CT differentiation during limb development82, a mechanism regulated by mechanical activity51. Similar regulation could occur during development, as CXCL12 induces CTGF expression in fibroblasts71 and TGF-β has been shown to cooperate with CXCL12 in carcinoma-associated fibroblasts to induce α-SMA expression83. One possibility could be that TGF-β, dowstream of mechanical signals, interacts with CXCL12 to regulate ICT-associated genes in the limb, among which CTGF and OSRs.
Our study demonstrates for the first time that CXCL12 and CXCL14 chemokines differentially regulate the expression of specific CT genes, participating to the orchestration of ICT, MCT and dense regular CT differentiation during chick limb development (Supp. Figure 4). The developmental role for CXCL12 and CXCL14 chemokines in CT differentiation is fully consistent with their recognised functions in adult fibrosis processes, such as in chronic fibrotic pathologies69,70,73,74 and cancer-induced fibrosis84. CXCL12 and CXCL14 regulate the expression of CT-associated transcription factors in chick limb and are themselves controlled by the anti-fibrotic factor BMP and by the fibrotic properties of mechanical forces of the musculoskeletal system. Such an unexpected role of these chemokines during CT differentiation can contribute to a better understanding of the fibrosis mechanisms in adult pathological conditions.
Methods
Chick embryos
Fertilized chick eggs from commercial sources (JA 57 strain, Institut de Sélection Animale, Lyon, France, and White Leghorn, HAAS, Strasbourg) were incubated at 38 °C in a humidified incubator until appropriate stages. Embryos were staged according to the number of days in ovo (E). All experiments on chick live embryos have been realized before E14 and consequently are not submitted to a licensing committee, in accordance to the European guidelines and regulations.
Constructs
The chicken CXCL12, CXCL14 and dnCXCR7 coding regions were amplified by PCR from an RT-PCR-derived cDNA library made from E5 chick limb, using primers containing ClaI restriction site. The dnCXCR7 coding region is a truncated form of CXCR7 lacking the C-terminal part of the sequence (Ray et al., 2012). CXCL12, CXCL14 and dnCXCR7 amplified sequence were then inserted into pCR-II TOPO vector using TOPO-TA cloning kit (Invitrogen). Inserted sequences were excised by digestion with ClaI and inserted into the ClaI site of replication-competent retroviral vector RCASBP(A)85. Clones containing the CXCL12, CXCL14 or dnCXCR7 coding regions in the sense orientation were selected. Mouse RCAS-mBmp4 and chick RCAS-cNOGGIN constructs were described previously86,87.
Production and grafting of recombinant/RCAS-expressing or control RCAS-expressing cells
Cells expressing RCAS-cCXCL12, RCAS-cCXCL14, RCAS-mBmp4 or RCAS-cNOGGIN and control cells were prepared for grafting as previously described54,88. Primary fibroblasts from E10 chick embryos were plated in vitro, transfected with the RCAS constructs and grown until 90% confluence. Transfected fibroblasts were then transferred to uncoated culture dishes to allow cell pellet formation before grafting. Pellets of approximately 50 μm in diameter were grafted into the right wing bud of E4 chick embryos. Embryos were harvested at various times after grafting and processed for in situ hybridization or immunohistochemistry to tissue sections. The left wing was used as an internal control. Owing to certain variability in the virus spread among embryos, the expression of the ectopic gene was systematically checked by in situ hybridization.
Lateral plate mesoderm electroporation
E2.5 chick embryos were electroporated as previously described50. PT2AL-CMV-TOMATO- T2A-CXCL12 (1,5–2 μg/μl) construct was mixed with the transposase vector CMV-T2TP (molar ratio 1/3) to allow stable integration of genes in the chick genome, in a solution containing 0.33% carboxymethyl cellulose, 1% Fast green, 1 mM MgCl2 in PBS. DNA mix was injected with a glass capillary in the coelomic cavity between somatopleural and splanchnopleural mesoderm, at the level of the forelimb territory. Homemade platinum electrodes were placed above and bellow the embryo, with the negative electrode inserted into the yolk and the positive electrode localized above the presumptive forelimb region. Electroporation was delivered using a Nepagene NEPA21 electroporator with the following parameters: 2 pulses of 70 V, 1ms duration with 100 ms interpulse interval followed by 5 pulses of 40 V, 2ms duration with 500 ms interpulse interval. Electroporated embryos were harvested 6 days later and indian ink was injected into the heart using a glass capillary, in order to stain vessel organization.
Bead implantation in chick limb buds
Heparin beads (Sigma) were soaked in 1 mg/ml of recombinant human FGF4 (R&D Systems) for 30 min on ice. FGF4 beads were grafted into the right wings of chick embryos at E4.5 and embryos were harvested 24 or 48 hours after grafting. Grafted right and contralateral left limbs were processed for in situ hybridization to sections.
Drug administration in ovo
The stock solution for decamethonium bromide (DMB, Sigma D1260) was prepared at 10% dilution in Hank’s solution (Sigma H9269). The DMB solution was freshly prepared before each experiment at 0.5% in Hank’s solution with 1% of Penicillin-Streptomycin (Gibco, 15140). 100 µl of the DMB or control solution (Hank’s solution with 1% of Penicillin-Streptomycin) were administrated in ovo at E4 of development. Embryos were harvested 48 to 72 hours after and processed for in situ hybridization or quantitative real-time PCR.
In situ hybridization and immunostaining to tissue sections
For hybridization and immunostaining on sections, embryos were fixed in a 4% paraformaldehyde solution in PBS supplemented with 4% sucrose and 0.1 mM CaCl2, rinsed in PBS, embedded in a 15% sucrose solution, frozen in chilled isopentane before cryostat sectioning at 10–20 μm. Sections were collected on Superfrost/Plus slides (CML, France) and processed for immunolabelling or in situ hybridization as described previously45. For grafted embryos, grafted and control limbs from the same experimental embryo were positioned in the same orientation for transverse sectioning to allow comparison. For in situ hybridization, the following digoxigenin-labeled mRNA probes were used: chick OSR1 and OSR2 21, chick CXCL12 45, chick SCX 89, chick COL1a1 and COL3a1 (produced from EST clones from ARK Genomics), mouse Bmp4 54. Chick CXCL14 probe was produced from an RT-PCR-derived cDNA library made from E5 chick limb, using primers previously described31. For immunostaining, the following primary antibodies were used: mouse monoclonal anti-MF20 (Developmental Studies Hybridoma Bank, non-diluted supernatant), rabbit polyclonal anti-PDGFRα (Santa Cruz, sc-338, 1/500 dilution), rabbit polyclonal anti-TCF4 (2569, Cell signalling, 1/100 dilution). DAPI (Sigma) was diluted at 1/1000. Slides were observed with a Nikon microscope and images collected with the QCapture Pro software (QImaging) and processed using Adobe Photoshop software.
RNA isolation, reverse transcription and quantitative real-time PCR
Total RNAs were extracted from chick limbs. 500ng to 1 µg RNAs were reverse-transcribed using the High Capacity Retrotranscription kit (Applied Biosystems). RT-qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems). Primer sequences used for RT-qPCR are listed in Table S1. The relative mRNA levels were calculated using the 2−ΔΔCt method90. The ΔCts were obtained from Ct normalized with chick GAPDH or S17 levels in each sample. For in vitro experiments, six cultures were used as independent RNA samples. For in vivo experiments, four forelimbs were used as independent RNA samples. Each sample was analysed in duplicate. Results were expressed as Standard Deviation (SD). Data were analysed by paired student t-test.
References
Omelyanenko, N. P. & Slutsky, L. I. Connective tissue: histophysiology, biochemistry, molecular biology. (CRC Press, Taylor and Francis Group, 2013).
Wynn, T. A. & Ramalingam, T. R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–40 (2012).
Chevallier, a, Kieny, M. & Mauger, A. Limb-somite relationship: origin of the limb musculature. J. Embryol. Exp. Morphol. 41, 245–258 (1977).
Wachtler, F., Christ, B. & Jacob, H. J. On the determination of mesodermal tissues in the avian embryonic wing bud. Anat. Embryol. (Berl). 161, 283–289 (1981).
Ordahl, C. P. & Le Douarin, N. M. Two myogenic lineages within the developing somite. Development 114, 339–53 (1992).
Grim, M. & Wachtler, F. Muscle morphogenesis in the absence of myogenic cells. Anat Embryol 183, 67–70 (1991).
Michaud, J. L., Lapointe, F. & Le Douarin, N. M. The dorsoventral polarity of the presumptive limb is determined by signals produced by the somites and by the lateral somatopleure. Development 124, 1453–63 (1997).
Chevallier, A. & Kieny, M. On the role of the connective tissue in the patterning of the chick limb musculature. Wilhelm Roux’s Arch. Dev. Biol. 191, 277–280 (1982).
Kieny, M. & Mauger, A. Immunofluorescent Localization of Extracellular Matrix Components During Muscle Morphogenesis. 1. In Normal Chick Embryos. J. Exp. Zool. 232, 327–341 (1984).
Zhang, G. et al. Development of tendon structure and function: Regulation of collagen fibrillogenesis. J. Musculoskelet. Neuronal Interact. 5, 5–21 (2005).
Gara, S. K. et al. Differential and restricted expression of novel collagen VI chains in mouse. Matrix Biol. 30, 248–257 (2011).
Schweitzer, R. et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855–3866 (2001).
Murchison, N. D. et al. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697–2708 (2007).
Léjard, V. et al. Scleraxis and NFATc regulate the expression of the pro-α1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 282, 17665–17675 (2007).
Kardon, G., Harfe, B. D. & Tabin, C. J. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev. Cell 5, 937–944 (2003).
Mathew, S. J. et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 138, 371–384 (2011).
Hasson, P. et al. Tbx4 and Tbx5 Acting in Connective Tissue Are Required for Limb Muscle and Tendon Patterning. Dev. Cell 18, 148–156 (2010).
Bagchi, R. A. et al. The transcription factor scleraxis is a critical regulator of cardiac fibroblast phenotype. BMC Biol. 14, 21 (2016).
Contreras, O., Rebolledo, D. L., Oyarzún, J. E., Olguín, H. C. & Brandan, E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell Tissue Res. 364, 647–660 (2016).
Hasson, P., Buono, J. D. & Logan, M. P. O. Tbx5 is dispensable for forelimb outgrowth. Development 134, 85–92 (2007).
Stricker, S., Brieske, N., Haupt, J. & Mundlos, S. Comparative expression pattern of Odd-skipped related genes Osr1 and Osr2 in chick embryonic development. Gene Expr. Patterns 6, 826–34 (2006).
Stricker, S. et al. Odd-skipped related genes regulate differentiation of embryonic limb mesenchyme and bone marrow mesenchymal stromal cells. Stem Cells Dev. 21, 623–33 (2012).
Bonafede, A., Köhler, T., Rodriguez-Niedenführ, M. & Brand-Saberi, B. BMPs restrict the position of premuscle masses in the limb buds by influencing Tcf4 expression. Dev. Biol. 299, 330–344 (2006).
Edom-Vovard, F., Schuler, B., Bonnin, M.-A., Teillet, M.-A. & Duprez, D. Fgf4 positively regulates scleraxis and tenascin expression in chick limb tendons. Dev. Biol. 247, 351–366 (2002).
Lejard, V. et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem. 286, 5855–5867 (2011).
Jenkins, R. H. & Fraser, D. J. BMP-6 emerges as a potential major regulator of fibrosis in the kidney. Am. J. Pathol. 178, 964–965 (2011).
Kim, K. K., Sisson, T. H. & Horowitz, J. C. Fibroblast Growth Factors and Pulmonary Fibrosis: It’s more complex than it sounds. J. Pathol. 6–9, https://doi.org/10.1002/path.4825 (2016).
Wynn, T. Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199–210 (2008).
Vasyutina, E. et al. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 19, 2187–98 (2005).
García-Andrés, C. & Torres, M. Comparative expression pattern analysis of the highly conserved chemokines SDF1 and CXCL14 during amniote embryonic development. Dev. Dyn. 239, 2769–77 (2010).
Gordon, C. T., Wade, C., Brinas, I. & Farlie, P. G. CXCL14 expression during chick embryonic development. Int. J. Dev. Biol. 55, 335–40 (2011).
Nagasawa, T. et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382, 635–638 (1996).
Zou, Y. R., Kottmann, A. H., Kuroda, M., Taniuchi, I. & Littman, D. R. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 393, 595–599 (1998).
Klein, R. S. et al. SDF-1 α induces chemotaxis and enhances Sonic hedgehog-induced proliferation of cerebellar granule cells. Development 128, 1971–1981 (2001).
Bagri, A. et al. The chemokine SDF1 regulates migration of dentate granule cells. Development 129, 4249–4260 (2002).
Molyneaux, Ka The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130, 4279–4286 (2003).
Boldajipour, B. et al. Control of chemokine-guided cell migration by ligand sequestration. Cell 132, 463–73 (2008).
Puchert, M. & Engele, J. The peculiarities of the SDF-1/CXCL12 system: in some cells, CXCR4 and CXCR7 sing solos, in others, they sing duets. Cell Tissue Res. 355, 239–53 (2014).
Hara, T. & Tanegashima, K. Pleiotropic functions of the CXC-type chemokine CXCL14 in mammals. J. Biochem. 151, 469–76 (2012).
Izukuri, K. et al. Chemokine CXCL14/BRAK transgenic mice suppress growth of carcinoma cell xenografts. Transgenic Res. 19, 1109–1117 (2010).
Tessema, M. et al. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene 29, 5159–70 (2010).
Tanegashima, K. et al. CXCL14 is a natural inhibitor of the CXCL12-CXCR4 signaling axis. FEBS Lett. 587, 1731–5 (2013).
Orr-Urtreger, A., Bedford, M. T., Do, M. S., Eisenbach, L. & Lonai, P. Developmental expression of the alpha receptor for platelet-derived growth factor, which is deleted in the embryonic lethal Patch mutation. Development 115, 289–303 (1992).
Tozer, S. et al. Involvement of vessels and PDGFB in muscle splitting during chick limb development. Development 134, 2579–2591 (2007).
Escot, S., Blavet, C., Härtle, S., Duband, J.-L. & Fournier-Thibault, C. Misregulation of SDF1-CXCR4 signaling impairs early cardiac neural crest cell migration leading to conotruncal defects. Circ. Res. 113, 505–16 (2013).
Escot, S. et al. Disruption of CXCR4 signaling in pharyngeal neural crest cells causes DiGeorge syndrome-like malformations. Development 143, 582–588 (2016).
Murdoch, C., Monk, P. N. & Finn, A. Cxc Chemokine Receptor Expression on Human Endothelial Cells. Cytokine 11, 704–712 (1999).
Tachibana, K. et al. The chemokine receptor CXCR4 is essential for vascularization of the gastrointestinal tract. Nature 393, 591–594 (1998).
Li, M. & Ransohoff, R. M. The roles of chemokine CXCL12 in embryonic and brain tumor angiogenesis. Semin. Cancer Biol. 19, 111–115 (2009).
Bourgeois, A. et al. Stable and bicistronic expression of two genes in somite- and lateral plate-derived tissues to study chick limb development. BMC Dev. Biol. 15, 39 (2015).
Havis, E. et al. TGFβ and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development 143, 3839–3851 (2016).
Weiskirchen, R. et al. BMP-7 as antagonist of organ fibrosis. Front. Biosci. 14, 4992–5012 (2009).
Schweitzer, R., Zelzer, E. & Volk, T. Connecting muscles to tendons: tendons and musculoskeletal development in flies and vertebrates. Development 137, 2807–2817 (2010).
Wang, H., Noulet, F., Edom-Vovard, F., Le Grand, F. & Duprez, D. Bmp Signaling at the Tips of Skeletal Muscles Regulates the Number of Fetal Muscle Progenitors and Satellite Cells during Development. Dev. Cell 18, 643–654 (2010).
Shwartz, Y., Blitz, E. & Zelzer, E. One load to rule them all: Mechanical control of the musculoskeletal system in development and aging. Differentiation 86, 104–111 (2013).
Esteves de Lima, J. et al. Specific pattern of cell cycle during limb fetal myogenesis. Dev. Biol. 392, 308–323 (2014).
Carver, W. & Goldsmith, E. C. Regulation of tissue fibrosis by the biomechanical environment. Biomed Res. Int. 2013, (2013).
Huang, A. H. et al. Musculoskeletal integration at the wrist underlies the modular development of limb tendons. Development 142, 2431–41 (2015).
Nowlan, N. C., Sharpe, J., Roddy, K. A., Prendergast, P. J. & Murphy, P. Mechanobiology of embryonic skeletal development: Insights from animal models. Birth Defects Res. Part C - Embryo Today Rev. 90, 203–213 (2010).
Pitsillides, A. A. Early effects of embryonic movement: ‘A shot out of the dark’. Journal of Anatomy 208, 417–431 (2006).
Landmesser, L. & Morris, D. G. The development of functional innervation in the hind limb of the chick embryo. J. Physiol. 249, 301–26 (1975).
Fredette, B. J. & Landmesser, L. T. A reevaluation of the role of innervation in primary and secondary myogenesis in developing chick muscle. Dev. Biol. 143, 19–35 (1991).
Sahin, H. & Wasmuth, H. E. Chemokines in tissue fibrosis. Biochim. Biophys. Acta - Mol. Basis Dis. 1832, 1041–1048 (2013).
Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 16, 582–598 (2016).
Wells, R. G. Tissue mechanics and fibrosis. 1832, 884–890 (2013).
Hamdan, R., Zhou, Z. & Kleinerman, E. S. SDF-1α induces PDGF- B expression and the differentiation of bone marrow cells into pericytes. Mol Cancer Res 9, 1462–1470 (2011).
Betsholtz, C. & Raines, E. W. Platelet-derived growth factor: a key regulator of connective tissue cells in embryogenesis and pathogenesis. Kidney Int. 51, 1361–9 (1997).
Horikawa, S. et al. PDGFRα plays a crucial role in connective tissue remodeling. Sci. Rep. 5, 17948 (2016).
Tan, J. et al. Loss of Twist1 in the Mesenchymal Compartment Promotes Increased Fibrosis in Experimental Lung Injury by Enhanced Expression of CXCL12. J. Immunol. 198, 2269–2285 (2017).
Neesse, A. & Ellenrieder, V. NEMO-CXCL12/CXCR4 axis: a novel vantage point for antifibrotic therapies in chronic pancreatitis? Gut 66, 211–212 (2017).
Lin, C. H. et al. CXCL12 induces connective tissue growth factor expression in human lung fibroblasts through the Rac1/ERK, JNK, and AP-1 pathways. PLoS One 9, 1–16 (2014).
Rodriguez-Nieves, J. A., Patalano, S. C., Almanza, D., Gharaee-Kermani, M. & Macoska, J. A. CXCL12/CXCR4 axis activation mediates prostate myofibroblast phenoconversion through non-canonical EGFR/MEK/ERK signaling. PLoS One 11, 1–14 (2016).
Jia, G. et al. CXCL14 is a candidate biomarker for Hedgehog signalling in idiopathic pulmonary fibrosis. Thorax 10.113 (2017).
Chen, L. et al. Overexpression of CXC chemokine ligand 14 exacerbates collagen-induced arthritis. J. Immunol. 184, 4455–4459 (2010).
Augsten, M. et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi-modal stimulator of prostate tumor growth. Proc. Natl. Acad. Sci. USA 106, 3414–3419 (2009).
Havis, E. et al. Transcriptomic analysis of mouse limb tendon cells during development. Development 141, 3683–96 (2014).
Khurana, S. et al. SMAD signaling regulates CXCL12 expression in the bone marrow niche, affecting homing and mobilization of hematopoietic progenitors. Stem Cells 32, 3012–3022 (2014).
Wells, R. G. & Discher, D. E. Matrix Elasticity, Cytoskeletal Tension, and TGF-β: The Insoluble and Soluble Meet. Sci. Signal. 1, 1–6 (2009).
Scott, A. et al. Mechanical force modulates scleraxis expression in bioartificial tendons. J. Musculoskelet. Neuronal Interact. 11, 124–132 (2011).
Chen, X. et al. Force and scleraxis synergistically promote the commitment of human ES cells derived MSCs to tenocytes. Sci. Rep. 2, 977 (2012).
Heinemeier, K. M. et al. Expression of collagen and related growth factors in rat tendon and skeletal muscle in response to specific contraction types. J. Physiol. 582, 1303–16 (2007).
Pryce, B. A. et al. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development 136, 1351–61 (2009).
Kojima, Y. et al. Autocrine TGF- and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc. Natl. Acad. Sci. 107, 20009–20014 (2010).
Allinen, M. et al. Molecular characterization of the tumor microenvironment in breast cancer. 6, 17–32 (2004).
Hughes, S. H., Greenhouse, J. J., Petropoulos, C. J. & Sutrave, P. Adaptor plasmids simplify the insertion of foreign DNA into helper-independent retroviral vectors. J. Virol. 61, 3004–12 (1987).
Duprez, D. et al. Overexpression of BMP-2 and BMP-4 alters the size and shape of developing skeletal elements in the chick limb. Mech. Dev. 57, 145–157 (1996).
Edom-Vovard, F., Bonnin, M. A. & Duprez, D. Misexpression of Fgf-4 in the chick limb inhibits myogenesis by down-regulating Frek expression. Dev. Biol. 233, 56–71 (2001).
Delfini, M. C., Hirsinger, E., Pourquié, O. & Duprez, D. Delta 1-activated notch inhibits muscle differentiation without affecting Myf5 and Pax3 expression in chick limb myogenesis. Development 127, 5213–5224 (2000).
Bonnin, M. A. et al. Six1 is not involved in limb tendon development, but is expressed in limb connective tissue under Shh regulation. Mech. Dev. 122, 573–585 (2005).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and. Methods 25, 402–408 (2001).
Acknowledgements
This work was supported by the Fondation pour la Recherche Médicale (FRM) DEQ. 20140329500, Agence Nationale de la Recherche (ANR) ANR-12-BSV1-0038, Association Française contre les Myopathies (AFM) N◦16826. SN was part of the MyoGrad International Research Training Group for Myology and received financial support from the AFM (AFM 20150532272).
Author information
Authors and Affiliations
Contributions
D.D. and C.F.T. conceived the experiments, S.N., C.B. and M.A.B. conducted the experiments; S.N., D.D. and C.F.T. analyzed the experiments; S.N., S.S., D.D. and C.F.T. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nassari, S., Blavet, C., Bonnin, MA. et al. The chemokines CXCL12 and CXCL14 differentially regulate connective tissue markers during limb development. Sci Rep 7, 17279 (2017). https://doi.org/10.1038/s41598-017-17490-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-17490-z
This article is cited by
-
Unexpected contribution of fibroblasts to muscle lineage as a mechanism for limb muscle patterning
Nature Communications (2021)
-
Evolution of the muscular system in tetrapod limbs
Zoological Letters (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.