Abstract
Ly6Chi inflammatory monocytes (iMO) are critical for host defense against toxoplasmosis and malaria but their role in leishmaniasis is unclear. In this study, we report a detrimental role of Ly6Chi iMOs in visceral leishmaniasis (VL) caused by Leishmania donovani. We demonstrate that Ly6Chi iMOs are continuously recruited into the spleen and liver during L. donovani infection and they are preferential targets for the parasite. Using microarray-based gene expression profiling, we show that Ly6Chi iMOs isolated from the infected liver and spleen have distinct phenotypic and activation profiles. Furthermore, we demonstrate that blocking the recruitment of Ly6Chi iMOs into the liver and spleen during L. donovani infection using a CCR2 antagonist reduces the frequency of the pathogenic IFN-γ/IL10 dual producer CD4+ T cells in the spleen and leads to a significant reduction in parasite loads in the liver and spleen. Using STAT1−/− mice we show that STAT1 is critical for mediating the recruitment of Ly6Chi iMOs into organs during L. donovani infection, and adaptive transfer of wild type Ly6Chi iMOs into STAT1−/− recipients renders them susceptible to disease. Our findings reveal an unexpected pathogenic role for Ly6Chi iMOs in promoting parasite survival in VL and open the possibility of targeting this population for host-directed therapy during VL.
Similar content being viewed by others
Introduction
Visceral leishmaniasis (VL) is the second leading cause of mortality among tropical diseases. It is caused by Leishmania donovani in Asia and Africa and L. infantum in South America and the Mediterranean basin. L. donovani is an obligate intracellular protozoan that infects professional phagocytes such as neutrophils, macrophages, dendritic cells, and spreads systemically infecting the spleen, liver and bone marrow1. As of yet, there is no vaccine available, and current drug treatments have adverse side effects, such as liver and pancreatic damage, and in some cases, death. Investigating the mechanisms that L. donovani exploits in order to survive in the host is highly relevant so as to understand disease progression and design new therapies.
It is well established that protective immunity against VL involves a Th1-associated response characterized by the IL12-IFNγ-iNOS pathway, which is necessary for optimal macrophage activation and formation of granulomas2,3. STAT1 is an important transcription factor that mediates IFNγ signaling and is central to the development of a Th1 immune response. Surprisingly, mice that are genetically deficient in STAT1, although unable to mount a Th1 immune response, are highly resistant to VL4. Mechanisms surrounding STAT1 mediated susceptibility to VL caused by L. donovani have remained incompletely understood, but previous studies suggest that T cell independent mechanisms are involved. Understanding the cellular mechanisms surrounding the resistance of STAT1−/− mice to VL will provide insights into L. donovani pathogenesis in the host and provide new strategies for treatment against VL.
At the cellular level, L. donovani preferentially infects Kupffer cells in the liver and marginal zone macrophages in the spleen5. Newly recruited macrophages which are derived from circulating monocytes are also potential host cells and play a crucial role in the establishment of L. donovani infection. These circulating monocytes are divided into two subsets: patrolling monocytes (characterized as CD11b+ CX3CR1lo Ly6Clow CCR2- CD115+) and inflammatory monocytes iMOs (characterized as CD11b+ CX3CR1lo Ly6Chi CCR2+ CD115+)6. iMOs exit the bone marrow in a CCR2 dependent manner, and once in circulation, they infiltrate the tissue3. iMOs are activated after pathogen recognition through Toll like receptors (TLRs) and/or IFNγ signaling and produce nitric oxide as well as inflammatory cytokines such as TNFα. Finally, iMOs differentiate into macrophages or dendritic cells (TipDCs) with enhanced microbicidal activity and are indispensable for resistance to a variety of pathogens7. Since the activation and subsequent differentiation of iMOs depends in part on STAT1 mediated IFNγ signaling, it is essential that we understand the role of this immune cell subset during infection with L. donovani.
iMOs play an important role in immunity against intracellular pathogens8. They have been shown to drive strong Th1 immune responses and protection against murine Toxoplasma infection9,10. Further, blockade of iMOs migration from the bone marrow to infected sites results in increased susceptibility to Listeria monocytogenes in mice3. iMOs are associated with protection against Plasmodium vivax 11, although they also contribute to brain pathology in a mouse model of experimental cerebral malaria. In Leishmania major infection, iMOs have also been shown to play a protective role12. In this study, we investigated the role of iMOs during VL. In contrast to most intracellular pathogens, our results reveal a detrimental role for this immune cell subset. Our results also provide an explanation for the previously observed surprising resistance to L. donovani infection that is observed in STAT1−/− mice.
Results
Dynamics of inflammatory monocyte recruitment during VL
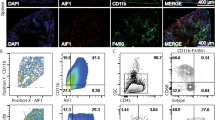
A variety of infections cause the recruitment of iMOs into infected tissues which results in pathogen clearance13,14. During murine VL, monocytes are recruited into the liver resulting in the formation of granulomas during early and chronic phase of the infection. However, what is not known is the kinetics of such monocyte recruitment, and whether the infiltrate consists of Ly6Chi iMOs. In order to investigate the early phase of the immune response during VL, we studied the mobilization of iMOs into L. donovani infected organs. To this end, we identified iMOs as CD11b+ Ly6Chi Ly6G− by flow cytometry. The proportion of iMOs increased after 24h post infection (pi) in the liver and spleen (Fig. 1A). Next, we investigated the kinetics of iMO recruitment. We found constant accumulation of iMOs in the liver and spleen over time reaching up to 15–25% of myeloid cells in both organs after 30 days of post infection (dpi) (Fig. 1B). We next investigated the ability of iMOs to phagocytize L. donovani parasites. Using transgenic DsRED–L. donovani parasites, we looked for infected cells after 24h of inoculation in the peritoneal cavity. We found three subsets infected with L. donovani. The cells harboring more parasites were Ly6Chi Ly6G−CD11b+. The second population of infected cells consisted of Ly6C+ Ly6G+ CD11b+ neutrophils. Interestingly, resident Ly6C-Ly6G-CD11b+ macrophages were also infected, however they represented the least infected cells among the three populations (Fig. 1C–F). Taken together, L. donovani infection resulted in early and continuous recruitment of iMOs into liver and spleen with high capability to uptake Leishmania parasites.
Leishmania donovani infection induced preferential recruitment of inflammatory monocytes. (A) Flow cytometric analysis of CD11b+ gated cells from the spleen and liver of L. donovani infected mice at 24 hr post infection. (B) Kinetics of inflammatory monocyte recruitment was evaluated by flow cytometry at the indicated time points in spleen and liver. Data represents the mean ± SEM from 1 of 2 independent experiments (n = 5/time point). L donovani-DsRED parasites were inoculated i.p. into WT mice, 24 h later peritoneal exudates were evaluated by Imaging-flow cytometry. Infected cells were gated as shown in (C) and analyzed for Ly6C and Ly6G expression (D). (E) Percentages of different subsets of infected cells, MO: Ly6Chi Ly6G-CD11b+, PMN: Ly6C+ Ly6G+ CD11b+, and Dneg: Ly6C-Ly6G-CD11bhi+ (n = 3). (F) Representative pictures of the different subsets infected with L. donovani analyzed by AMNIS Flow sight.
Characterization of inflammatory monocytes recruited during L. donovani infection
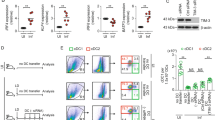
In order to confirm the identity of CD11b+ Ly6ChiLy6G- cells observed during L. donovani infection, we examined the expression of CD115 (MCSF receptor) and the hallmark chemokine receptor CCR23. These cells expressed CD115 and CCR2 on their cell surface, which is in agreement with the phenotype described for inflammatory monocytes15, (Fig. 2A). In some infections, inflammatory monocytes gain CD11c expression and are known as TipDCs16,17. However, during chronic infection with L. donovani, iMOs did not upregulate CD11c, but expressed intermediate levels of MHCII and F4/80 on their membrane (Fig. 2A), which is suggestive of their differentiation into the macrophage lineage. iMOs are known to express high levels of iNOS in several infections, and nitric oxide production is important in L. donovani resistance18. Interestingly iNOS expression was higher in liver than in the spleen after 30 dpi, and a similar trend was found for TNFα expression. Unlike iNOS, arginase is associated with Leishmania survival inside macrophages19. iMOs isolated from the spleen at 30 dpi showed higher arginase expression compared to liver isolated iMOs (Fig. 2B). Interestingly, arginase/iNOS ratio was higher in the spleen where the infection turns progressive, and the opposite trend was found in the liver where the infection starts resolving after 30 dpi (Fig. 2C). It appears from these data that, unlike the liver, the splenic microenvironment causes differentiation of iMOs into a phenotype that favors Leishmania survival. Recently, it was reported that iMOs can produce PGE2 depending on the gut microbiota20. COX2, an upstream regulator of PGE2 production was slightly overexpressed by iMOs in both the liver and the spleen (Fig. 2B)
Characterization of inflammatory monocytes phenotype during Leishmania donovani infection. (A) Flow cytometric analysis of Ly6ChiLy6G-CD11b+ cells from uninfected and L. donovani infected WT mice 30 days post infection. Cells were gated on CD11b+ and analyzed for monocyte/macrophages/DC associated markers. Representative overlaid histograms plots were shown. (B) Gene expression analysis of sorted Ly6ChiLy6G-CD11b+ cells from spleen and liver of L. donovani infected mice. Data represents fold change relative to sorted cells from uninfected mice. Bar graphs show the average gene expression of pooled Ly6ChiLy6G-CD11b+ cells sorted from infected mice (n = 4). (C) Gene expression ratio of Arginase:iNOS from Ly6Chi Ly6G- CD11b+ cells sorted from livers and spleens.
Inflammatory monocytes isolated from spleen and liver during VL present differential activation state
The uncommon arginase expression in monocytes isolated from spleen (Fig. 2C) lead us to expand the characterization of those iMOs by using microarray based gene expression profiling. Among the top upregulated genes, we found S100A9, S100A8 and CXCL9 (Fig. 3F). S100A8/9 functions as a chemoattractant for monocytes and neutrophils, while CXCL9 attracts CXCR3 expressing cells such as T and NK cells. These chemokines are enhanced with IFN stimulation21. Also, we found upregulation of IFN signaling pathway by increased expression of IRF1 and IGR1. Interestingly, when we compared IRF1 and IRG1 we found higher expression in the liver than in the spleen, which is opposite compared with S100A8/9 expression (Fig. 3F). Further, pathway analyses using Ingenuity Pathway Analysis software revealed other characteristics of iMOs, such as up regulation of antigen presentation associated genes and interferon signaling and complement activation in splenic iMOs after infection (Fig. 3C). Interestingly, splenic iMOs showed enhanced ubiquitination pathway protein expression and enhanced expression of oxidative phosphorylation which has been associated with M2 macrophage metabolism (Fig. 3C,D)22, which is in line with our observation of splenic iMOs expressing elevated arginase transcripts (Fig. 2B). Upstream analysis revealed a role for IFNs, STAT1, TLRs, IL1B and IL-6 as main regulators for iMOs gene expression (Fig. 3A,B). To identify key molecules involved in iMO phenotype, we generated a pathway for iMO, interestingly; STAT1 pointed was a key player in iMOs phenotype (Fig. 3E).
STAT1 is a key molecule in iMOs phenotype. Ly6Chi Ly6G-CD11b+ cells were sorted from the pooled spleen suspensions of naïve or 30 dpi mice (n = 4). RNA obtained from sorted cells was analyzed in Affymetrix microarray chip. Data was analyzed with Ingenuity pathways analysis software, and upstream regulators and networks were constructed. (A) Top upstream cytokine regulators on infected iMOs. (B) Top transcription factors found by IPA analysis. (C) Top canonical pathways associated with iMOs infection. (D) Network of genes involved in top 10 upstream regulators. (E) STAT1 network in infected iMOs. (F) Validation of up regulated genes by RT-PCR, represented fold increase is compared to WT un-infected controls. Bar graphs show the average gene expression of pooled Ly6ChiLy6G-CD11b+ cells sorted from infected mice (n = 4).
STAT1 is necessary for iMO accumulation in spleen and liver during L. donovani infection
The central roles of STAT1 in the genome profile of iMOs lead us to investigate them in STAT1KO mice. Previously we found that STAT1 KO mice were highly resistant to VL in a T cell independent fashion4. Some studies have shown that IFNs could be important for monocyte differentiation in an IRF dependent pathway23. The main IFNγ intracellular signaling adaptor is STAT1. However, the role STAT1 plays in monocyte accumulation is unknown. We first investigated the accumulation of iMOs in STAT1 KO mice. As showed before (Fig. 1A) WT mice had enhanced accumulation of CD11b+ Ly6ChiLy6G- cells. Interestingly, spleens and livers of L. donovani infected STAT1KO mice showed profound defect in Ly6C monocyte accumulation (Fig. 4A,B). We then hypothesized that the defect in monocyte recruitment may account for the ability of STAT1 KO mice to efficiently control L. donovani infection. To test this hypothesis, we transferred bone marrow Ly6Chi WT monocytes into STAT1 KO mice on 1 dpi. After 20 days of infection STAT1KO mice that received WT monocytes showed two times more parasites in the spleen than control STAT1 KO mice (Fig. 4C). However, we did not find differences in parasitic loads in the liver (data not shown). Splenocytes from STAT1 KO mice proliferated more than those from WT mice after Leishmania antigen re-stimulation. However, the proliferation of STAT1 KO splenocytes was slightly reduced by transferred WT iMOs (Fig. 4D). WT infected mice produced enhanced levels of IFNγ and IL-10 in splenocyte culture supernatants upon L. donovani Antigen (LdAg) stimulation. In contrast, the culture supernatants from STAT1 deficient mice showed reduced levels of IFNγ and IL-10 (Fig. 4G,H). Strikingly, we observed significantly higher levels of IL-17 production from STAT1KO mice compared to the WT mice (Fig. 4I). Interestingly, STAT1 KO mice that received WT monocytes recovered IL-10 production and showed impaired IL-17 production. In line with other reports showing a relationship between IL-17 levels and neutrophil accumulation we found significantly lower neutrophil frequencies in spleens and livers of STAT1 KO mice that received WT monocytes compared with their controls (Fig. 4E,F).
STAT1 plays a critical role in iMO accumulation in L. donovani infected tissues. (A) Flow cytometric analysis of Ly6Chi cells from the spleen of L. donovani infected WT and STAT1KO mice at 20 dpi. (B) Flow cytometric analysis of Ly6Chi cells from the liver of L. donovani infected WT and STAT1KO mice at 20 dpi. (C) Bone marrow iMOS were sorted from naïve WT mice and transferred into L. donovani infected STAT1KO mice at 24 h pi, Parasitic loads were analyzed at 20 dpi. (D) Splenocytes were re-stimulated with L. donovani antigen for 72 h, proliferation was avaluated by using Alamar Blue. (E,F) Percentages of Ly6Chi (Ly6Chi Ly6G- CD11b+), Ly6Clow (Ly6Clow Ly6G-CD11b+) and PMN (Ly6C+ Ly6G+ CD11b+) in spleen and liver respectively. (G–I) Supernatants from antigen re-stimulated splenocytes were evaluated for IFNγ, IL-10 and IL-17 production by ELISA. Data represented from 1 of 3 independent experiments. n = 5, *p < 0.05.
Blocking iMO accumulation decreases parasitic loads in liver and spleen
Our data showed that deficient recruitment of iMOs in STAT1KO mice improved resistance to VL. Monocytes egress from the bone marrow in a CCR2 dependent fashion3. In order to block monocyte migration into the infected tissues we administrated a CCR2 specific antagonist RS-504393, as was previously used in a fibrosis model24. We injected CCR2 antagonist once daily from day 1 to day 29 pi (CCR2A) and another group once daily from day 20 to day 30 pi (CCR2B). Both modes of RS-504393 administration effectively reduced the numbers of inflammatory monocytes in spleens and livers (Fig. 5A,E) without affecting the frequency of the CD115+ Ly6Clo population (Fig. 5B,F). These findings strongly suggest that iMOs accumulated during VL were bone marrow derived and their migration was CCR2 dependent. Interestingly, mice which received RS-504393 presented less splenomegaly and hepatomegaly compared with control mice (Fig. 5C,G). Importantly, blocking iMOs accumulation by CCR2 antagonist resulted in significantly decreased parasitic loads in spleens and livers (Fig. 5D,H). A previous study by Sato et al.25 had found genetically resistant C57BL/6 mice lacking CCR2 develop similar parasitic burdens compared to WT upon L. donovani infection. Although Sato et al. did not examine the impact of CCR2 deficiency on iMOs recruitment to organs, these observations suggest that CCR2 and possibly iMOs recruited to the organs may not play a critical role in immunity against L. donovani. In an independent experiment by using luciferin expressing parasites, we also confirmed that CCR2 antagonist treatment successfully reduced the parasitic loads in livers, spleens and lymph nodes of L. donovani infected mice (Fig. 5I,J). Additionally, livers from mice treated with RS-504393 showed no granuloma formation irrespective of the period of RS-504393 administration (Supplementary Figure 1).
Administration of CCR2 antagonist reduced monocyte recruitment and parasitic loads. L. donovani infected mice were treated with CCR2 antagonist RS-504393, once daily from day 1 to day 30 pi (CCR2A) or once daily from day 20 to day 30 pi (CCR2B) respectively. Flow cytometry analysis of frequencies of Ly6Chi cells in spleen (Fig. 5A) and liver (Fig. 5E). Flow cytometry analysis of frequencies of Ly6Clow cells in spleen (Fig. 5B) and liver (Fig. 5F). Data shown is from one of the three experiments (n = 4). Spleen weight (Fig. 5C) and liver weight (Fig. 5G) percentages of body weight were represented at 30 dpi. Combined data from two independent experiments N = 8, *p < 0.05 **p = 0.01 ***p = 0.001. (I,J) Representative images and Kinetics of Luciferin-expression L. donovani was analyzed by IVIS imaging system at the indicated time points. Data representative of the mean ± SEM of photons/second/cm2 (n = 3).
CCR2 inhibition decreased IFNγ IL-10 double producers CD4+ T cells
In order to investigate if the absence of iMOs affected the host immune response, we evaluated the cytokine profile of CCR2 inhibitor treated mice. We found that CCR2 inhibitor administration resulted in decreased IL-10 production by splenocytes independent of the time of administration of CCR2 antagonist (Fig. 6A). Interestingly, CCR2 inhibitor administration had no effects on IFNγ and slightly, but not significantly, decreased IL-4 production, leaving the Th1/Th2 response intact (Fig. 6C,B). IL-10 was significantly diminished when we administered CCR2 antagonist, correlating with disappearance of inflammatory monocytes (Fig. 5A,E). However, we did not observe up-regulation of IL-10 mRNA in inflammatory monocytes in the spleen (Fig. 2B), suggesting that iMO could be influencing other cells to produce IL-10. In order to evaluate the source of IL-10, we performed intracellular cytokine staining of splenocytes from chronically infected mice. We found a CD4+ T cell population positive for both IFNγ and IL-10 (Fig. 6D). This population has been reported to be responsible for non-healing lesions during L. major infection and with the progression of VL26,27. In our experiments, RS-504393 treated mice showed reduced numbers of IFNγ IL10 double producer CD4+ T cells (Fig. 6D,E) and correlated with decreased IL-10 production found in splenocyte supernatants (Fig. 6A). We also noticed that antigen specific splenocyte proliferation was enhanced in RS-504393 treated mice (Fig. 6H). In line with this, administration of CCR2 antagonist resulted in enhanced frequencies of CD4+ T cells (Fig. 6G) and comparable numbers of CD4+ IFNγ+ cells in the spleen (Fig. 6F). The splenocytes of CCR2 antagonist received mice also showed increased T cell proliferation upon L. donovani antigen stimulation (Fig. 6H).
CCR2 antagonist reduced percentages of IFNγ+ IL10+ double producer CD4 cells. Splenocytes were harvested from L. donovani infected, CCR2A, CCR2B and vehicle treated mice at 30 dpi, re-stimulated with LdAg for 72 h, and release of cytokines was evaluated in the culture supernatants. (A) IFNγ, (B) IL-4 and (C) IL-10 release was evaluated by ELISA. (D) Representative dot plots showing IFNγ and/or IL10 production by CD4+ gated cells. Analysis of (E) frequencies of IFNγ IL-10 double producers CD4+ cells in the spleen, (F) IFNγ CD4+ producers cells, and (G) total CD4+ cells n = 4. (H) Proliferation analysis by alamar blue of splenocytes stimulated 72 h with L. donovani antigens. n = 5, *p < 0.05 **p = 0.01.
Discussion
In contrast with other infectious diseases, we found here that iMOs accumulated during VL and served as a continuous source of host cells for L. donovani parasite infection. Early recruited iMOs during VL did not show the classical activation profile, and lacked iNOS, TNFα, as well as expression markers for dendritic cells. iMOs from chronic infection demonstrated polarized profiles towards an inflammatory phenotype (iNOS) in the liver and an anti-inflammatory phenotype (arginase) in the spleen. This suggests that early contact with L. donovani is not able to trigger iMO activation and their polarization requires further help from the adaptive immune response. In mice, L. donovani profoundly infects the liver in the first 3–4 weeks and then the infection is resolved via an IL-12-IFNγ-iNOS dependent axis18,28. In the spleen the infection is chronic, progressive and does not resolve. This correlates with the high levels of iNOS we observed in liver iMOs which may partly account for their ability to kill the parasites and resolve liver infection29. On the other hand, higher arginase expression in splenic iMOs could potentially facilitate parasite growth and avoid its elimination.
It must be noted that blocking iMO accumulation was sufficient to avoid parasite establishment in the liver and spleen. This suggests that iMOs promote VL primarily as initiators of L. donovani establishment. Supporting this idea, we showed that STAT1 was required for iMO accumulation. This explains the resistant phenotype of STAT1 KO mice even in the absence of IFNγ signaling. It appears from this data that although STAT1 signaling is generally protective against VL, in some cases it can be detrimental, and could cause the accumulation of host reservoir cells for L. donovani infection4. STAT1 may participate in the differentiation of monocytes since IFNs play a role in monopoiesis23. IFNs are also important for the upregulation of integrins that facilitate iMO migration through the endothelia and tissues and by enhancing chemokine expression30. However, the exact mechanism of STAT1-mediated monocyte accumulation during VL requires further research.
iMOs require TLR or IFNγ stimulation to become activated. In contrast, the presence of IL-10 impairs their differentiation into TipDCs31. The ability of L. donovani to subvert TLR and IFNγ signaling and enhance IFNγ/IL10 producer CD4+ T cells can explain the inability of iMOs to become fully activated during VL. The iMO phenotype found in VL was in striking contrast to that found during cutaneous leishmaniasis, where iMOs differentiated into TipDCs with high nitric oxide production and leishmanicidal activity32. It has been proposed that the tissue microenvironment may control the immune response33, but parasites can also directly modify the behavior of innate myeloid cells34. In our study we still do not understand what causes the differences observed in monocyte polarization in the liver and spleen during VL, but it is possible that tissue-derived factors may play a role in regulating iMO activation.
Our results demonstrate as a proof of concept that blocking iMOs results in reduced parasitic loads during L. donovani infection. Importantly the absence of iMOs negatively affected the numbers of IFNγ/IL10+ CD4+ T cells which are also dependent on the presence of CD11c+ DCs and play a detrimental role during VL27. Interestingly, blocking iMO recruitment reduced granuloma formation. Although the formation of granulomas is important for L. donovani elimination from the liver, it has been shown that L. donovani elimination can be achieved in absence of granulomas35. The high expression of CXCL9 and S100A8/9 may account for the ability of iMOs to recruit neutrophils, monocytes and T cells into the infected site and shape granuloma formation.
In conclusion, our data provide evidence of a novel mechanism by which L. donovani successfully establishes infection in the host, which relies on the recruitment of iMOs. In this experimental VL model, these cells are critical for sustaining L. donovani infection by functioning as a host cell reservoir for the parasite, in addition to modulating the host immune response. However, it must be noted that there are significant differences between the well-established experimental VL model used in this study and human VL, which could have an impact on the significance of our results to human disease. For one thing, the intravenous injection of L. donovani amastigotes into mice contrasts with the natural sand-fly mediated cutaneous route of transmission of L. donovani promastigotes in humans. This obviously affects the nature of local and systemic immune responses involved in immunity to the human disease. Nevertheless, our novel findings underscore the importance of a unique immune cell subset that mediates the establishment of L. donovani infection in mice, which can potentially be exploited in the prevention and treatment of VL.
Materials and Methods
Animals
STAT1−/− (C.129S-Stat1〈tm1Dlv〉) mice were generated as described previously4. 8–10 weeks old female BALB/c mice were purchased from Envigo (previously Harlan laboratories, Indianapolis, IN, USA). All experimental mice were maintained in vivarium at The Ohio State University in compliance with the guidelines of OSU ULAR. All experimental procedures were performed according to the approved experimental protocol 2010A0048-R2 by The Ohio State University institutional guidelines for animal research (OSU-IACUC).
Parasites and infection protocol
Leishmania donovani (LV82 strain) amastigotes were isolated from the spleen of previously infected Golden Syrian hamsters. Red fluorescent protein expressing DsRed2 transgenic L. donovani LV82 parasites were obtained from infected stock mice as described previously36. All experimental mice were infected with 107 L. donovani LV82 or DsRed2 LV82 amastigotes by tail vein injection. Groups constituted of three or five mice and were sacrificed at different time points post-infection for further analysis.
Parasite burden calculation
At different time points during the infection, livers and spleens were harvested, weighed, and impression smears were taken. Smears were stained with Giemsa stain (Sigma-Aldrich, St. Louis, MO, USA). Parasite loads were counted under a microscope by a certified pathologist. Parasitic loads were expressed as Leishmania donovani Units (LDU) = Number of amastigotes per 1000 nucleated cells × organ weight (grams).
Treatment of mice
To evaluate the role of CCR2 inflammatory monocytes, we administered a CCR2 antagonist RS-504393 (R&D systems, Minneapolis, MN, USA) intraperitoneally. One group of mice received RS-504393 (2 mg/kg i.p.) one day after infection daily for 29 days (CCR2 A), second group received RS-504393 (2 mg/kg i.p) daily from days 20–30 dpi (CCR2 B). Control mice were injected with PBS intraperitoneally.
Cytokine ELISA and Alamar blue proliferation assay
Splenocytes isolated from WT and STAT1−/− mice were plated at a concentration of 0.5 × 106 cells per well in quadruplicates in 96-well tissue culture plates (Thermo-Fisher Scientific, Waltham, MA, USA). Cells were then stimulated with 20 ug/ml of L. donovani antigen (LdAg) prepared by freeze-thawing L. donovani promastigotes. Supernatants were collected after 72 h of incubation and analyzed for the production of IFN-γ, IL-4, IL-6, IL-10, IL-13 and IL-17 by ELISA (all capture and detection antibodies were purchased from Biolegend, San Diego, CA, USA) and recombinant mouse standards were purchased from BD Biosciences (San Jose, CA, USA). For proliferation assays 10% Alamar blue (Life technologies, Carlsbad, CA) was added 48h after stimuli and readings were taken after additional 18 hours of incubation. Data represent fluorescence units calculated by subtracting the fluorescence of unstimulated controls versus L. donovani stimulated cells. ELISA plates were read at an absorbance of 405 nm, and for Alamar blue, plates were read at 570 and 600nm using Spectramax M3 microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA) and concentrations were determined by Softmax Pro software (Molecular Devices LLC, Sunnyvale, CA, USA).
Flow cytometry
Single cell suspensions from liver and spleen were collected gently extruding the tissues through a 70 uM cell strainer (BD Biosciences, San Jose, CA, USA). Blood was recovered from tail vein bleeding. Red blood cells were lysed with ACK lysis solution. One million cells were blocked with mouse serum and stained with different cocktail antibodies or their respective isotype controls for 20 minutes at 4 °C. Ly6C APC, Ly6G PE, CD11b FITC, CD62L PercP, MHCII PE, F480 PercypCy7, CD115 PercP, and iNOS PE were purchased from Biolegend (San Diego, CA, USA), and CCR2 Alexa 700 or FITC, Arginase APC from R&D systems (Minneapolis, MN, USA). For intracellular detection of iNOS and arginase, cell membrane was stained first and then fixed with 2% paraformaldehyde for 10 minutes. Cell permeabilization was carried out using Biolegend permeabilization buffer followed by 45-minute incubation with anti iNOS and anti-arginase antibodies. Flow cytometry was performed using by BD FACSCalibur (San Jose, CA, USA) or Flowsight AMNIS (Seattle, WA, USA) cytometers. Cell sorting was performed using a BD FACS Aria (San Jose, CA, USA). Sorted Ly6Chi Ly6G-CD11b+ cells were isolated from bone marrow of BALB/c naïve mice, and 1 × 106 Ly6C+ Ly6G-CD11b+ cells were transferred i.v. into STAT1 KO mice one day before Leishmania donovani infection. For monocyte and neutrophil functional assays, Ly6Chi Ly6G- CD11b+ or Ly6C+ Ly6G+ CD11b+ expressing cells were isolated from the spleens of naïve or infected mice. Data was analyzed using Flow jo software (FLOWJO, LLC, Ashland, OR, USA).
Real Time PCR
Total RNA was extracted from both the spleens and livers of experimental mice by TRIzol extraction method (purchased from Life technologies (Carlsbad, CA)), making sure to minimize any contaminating genomic DNA material. cDNA was prepared by iScript reverse transcriptase and RT PCR reactions were done by using IQ SYBR green super mix and a CFX 96 RT-PCR cycler (purchased from Bio-Rad, Hercules, CA, USA). Pre-validated primers were selected from the Primer bank website (http://pga.mgh.harvard.edu/primerbank) and are listed in Supplementary Table 1. Samples were run in technical triplicates, and appropriate positive and negative controls were included for each RT-PCR experiment. Data obtained were normalized using the housekeeping gene β-actin and presented as the fold induction over uninfected WT mice.
Microarray
Five ng of total RNA was amplified using the NuGen Ovation Pico WTA System V2 and labeled by using the NuGen Encore Biotin Module (San Carlos, CA, USA). The samples were then hybridized onto the Mouse Transcriptome arrays (MTA) for 16 hours at 45 °C. The samples were then washed and stained on the Affymetrix Fluidics 450 system according to manufacturer protocols and scanned on the Affymetrix GeneChip scanner 3000 7G (Sunnyvale, CA, USA).
Statistical analysis
Student’s unpaired t-test was used to determine statistical significance of differences in the values. A value of p < 0.05 was considered significant.
References
Gupta, G., Oghumu, S. & Satoskar, A. R. Mechanisms of immune evasion in leishmaniasis. Advances in applied microbiology 82, 155–184, https://doi.org/10.1016/b978-0-12-407679-2.00005-3 (2013).
McFarlane, E. et al. Neutrophils contribute to development of a protective immune response during onset of infection with Leishmania donovani. Infection and immunity 76, 532–541, https://doi.org/10.1128/iai.01388-07 (2008).
Serbina, N. V. & Pamer, E. G. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nature immunology 7, 311–317, https://doi.org/10.1038/ni1309 (2006).
Rosas, L. E. et al. Cutting edge: STAT1 and T-bet play distinct roles in determining outcome of visceral leishmaniasis caused by Leishmania donovani. Journal of immunology (Baltimore, Md.: 1950) 177, 22–25 (2006).
Jenkins, S. J. et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science (New York, N.Y.) 332, 1284–1288, https://doi.org/10.1126/science.1204351 (2011).
Geissmann, F., Jung, S. & Littman, D. R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 (2003).
Auffray, C., Sieweke, M. H. & Geissmann, F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annual review of immunology 27, 669–692, https://doi.org/10.1146/annurev.immunol.021908.132557 (2009).
Soudja, S. M., Ruiz, A. L., Marie, J. C. & Lauvau, G. Inflammatory monocytes activate memory CD8(+) T and innate NK lymphocytes independent of cognate antigen during microbial pathogen invasion. Immunity 37, 549–562, https://doi.org/10.1016/j.immuni.2012.05.029 (2012).
Biswas, A. et al. Ly6C(high) monocytes control cerebral toxoplasmosis. Journal of immunology (Baltimore, Md.: 1950) 194, 3223–3235, https://doi.org/10.4049/jimmunol.1402037 (2015).
Dunay, I. R. et al. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity 29, 306–317, https://doi.org/10.1016/j.immuni.2008.05.019 (2008).
Antonelli, L. R. et al. The CD14+CD16+ inflammatory monocyte subset displays increased mitochondrial activity and effector function during acute Plasmodium vivax malaria. PLoS pathogens 10, e1004393, https://doi.org/10.1371/journal.ppat.1004393 (2014).
Glennie, N. D., Volk, S. W. & Scott, P. Skin-resident CD4+ T cells protect against Leishmania major by recruiting and activating inflammatory monocytes. PLoS pathogens 13, e1006349, https://doi.org/10.1371/journal.ppat.1006349 (2017).
Dunay, I. R., Fuchs, A. & Sibley, L. D. Inflammatory monocytes but not neutrophils are necessary to control infection with Toxoplasma gondii in mice. Infection and immunity 78, 1564–1570, https://doi.org/10.1128/iai.00472-09 (2010).
Jia, T., Leiner, I., Dorothee, G., Brandl, K. & Pamer, E. G. MyD88 and Type I interferon receptor-mediated chemokine induction and monocyte recruitment during Listeria monocytogenes infection. Journal of immunology (Baltimore, Md.: 1950) 183, 1271–1278, https://doi.org/10.4049/jimmunol.0900460 (2009).
Geissmann, F. et al. Development of monocytes, macrophages, and dendritic cells. Science (New York, N.Y.) 327, 656–661, https://doi.org/10.1126/science.1178331 (2010).
Serbina, N. V., Salazar-Mather, T. P., Biron, C. A., Kuziel, W. A. & Pamer, E. G. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity 19, 59–70 (2003).
De Trez, C. et al. iNOS-producing inflammatory dendritic cells constitute the major infected cell type during the chronic Leishmania major infection phase of C57BL/6 resistant mice. PLoS pathogens 5, e1000494, https://doi.org/10.1371/journal.ppat.1000494 (2009).
Murray, H. W. & Nathan, C. F. Macrophage microbicidal mechanisms in vivo: reactive nitrogen versus oxygen intermediates in the killing of intracellular visceral Leishmania donovani. The Journal of experimental medicine 189, 741–746 (1999).
Iniesta, V. et al. Arginase I induction during Leishmania major infection mediates the development of disease. Infection and immunity 73, 6085–6090, https://doi.org/10.1128/iai.73.9.6085-6090.2005 (2005).
Grainger, J. R. et al. Inflammatory monocytes regulate pathologic responses to commensals during acute gastrointestinal infection. Nature medicine 19, 713–721, https://doi.org/10.1038/nm.3189 (2013).
Padovan, E., Spagnoli, G. C., Ferrantini, M. & Heberer, M. IFN-alpha2a induces IP-10/CXCL10 and MIG/CXCL9 production in monocyte-derived dendritic cells and enhances their capacity to attract and stimulate CD8+ effector T cells. Journal of leukocyte biology 71, 669–676 (2002).
Rodriguez-Prados, J. C. et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. Journal of immunology (Baltimore, Md.: 1950) 185, 605–614, https://doi.org/10.4049/jimmunol.0901698 (2010).
de Bruin, A. M. et al. IFNgamma induces monopoiesis and inhibits neutrophil development during inflammation. Blood 119, 1543–1554, https://doi.org/10.1182/blood-2011-07-367706 (2012).
Kitagawa, K. et al. Blockade of CCR2 ameliorates progressive fibrosis in kidney. The American journal of pathology 165, 237–246, https://doi.org/10.1016/s0002-9440(10)63292-0 (2004).
Sato, N. et al. Defects in the generation of IFN-gamma are overcome to control infection with Leishmania donovani in CC chemokine receptor (CCR) 5-, macrophage inflammatory protein-1 alpha-, or CCR2-deficient mice. Journal of immunology (Baltimore, Md.: 1950) 163, 5519–5525 (1999).
Owens, B. M. et al. IL-10-producing Th1 cells and disease progression are regulated by distinct CD11c(+) cell populations during visceral leishmaniasis. PLoS pathogens 8, e1002827, https://doi.org/10.1371/journal.ppat.1002827 (2012).
Anderson, C. F., Oukka, M., Kuchroo, V. J. & Sacks, D. CD4(+)CD25(−)Foxp3(−) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. The Journal of experimental medicine 204, 285–297, https://doi.org/10.1084/jem.20061886 (2007).
Oghumu, S. et al. STAT4 is critical for immunity but not for antileishmanial activity of antimonials in experimental visceral leishmaniasis. European journal of immunology 44, 450–459, https://doi.org/10.1002/eji.201343477 (2014).
Cervia, J. S., Rosen, H. & Murray, H. W. Effector role of blood monocytes in experimental visceral leishmaniasis. Infection and immunity 61, 1330–1333 (1993).
Issekutz, T. B. In vivo blood monocyte migration to acute inflammatory reactions, IL-1 alpha, TNF-alpha, IFN-gamma, and C5a utilizes LFA-1, Mac-1, and VLA-4. The relative importance of each integrin. Journal of immunology (Baltimore, Md.: 1950) 154, 6533–6540 (1995).
Guilliams, M. et al. IL-10 dampens TNF/inducible nitric oxide synthase-producing dendritic cell-mediated pathogenicity during parasitic infection. Journal of immunology (Baltimore, Md.: 1950) 182, 1107–1118 (2009).
Leon, B., Lopez-Bravo, M. & Ardavin, C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity 26, 519–531, https://doi.org/10.1016/j.immuni.2007.01.017 (2007).
Matzinger, P. & Kamala, T. Tissue-based class control: the other side of tolerance. Nature reviews. Immunology 11, 221–230, https://doi.org/10.1038/nri2940 (2011).
Terrazas, C. A., Terrazas, L. I. & Gomez-Garcia, L. Modulation of dendritic cell responses by parasites: a common strategy to survive. Journal of biomedicine & biotechnology 2010, 357106, https://doi.org/10.1155/2010/357106 (2010).
Murray, H. W. Tissue granuloma structure-function in experimental visceral leishmaniasis. International journal of experimental pathology 82, 249–267 (2001).
Lezama-Davila, C. M. et al. Leishmanicidal activity of two naphthoquinones against Leishmania donovani. Biological & pharmaceutical bulletin 35, 1761–1764 (2012).
Acknowledgements
Authors would like to thank Dr. Stephen M. Beverley for kindly providing us the red fluorescent protein expressing DsRed2 transgenic L. donovani LV82 parasites. This project was supported in part by an appointment to the Research Participation Program at the Food and Drug Administration (FDA), administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the Environmental Protection Agency.
Author information
Authors and Affiliations
Contributions
C.T., S.V., S.O., J.K., H.M.S., N.A. performed the experiments; S.V. estimated parasitic loads, L.Y. analyzed gene profile. S.B. and H.N. provided critical materials for the experiments, C.T., S.O. and A.S. conceived, designed the study and wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Terrazas, C., Varikuti, S., Oghumu, S. et al. Ly6Chi inflammatory monocytes promote susceptibility to Leishmania donovani infection. Sci Rep 7, 14693 (2017). https://doi.org/10.1038/s41598-017-14935-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-14935-3
This article is cited by
-
Macrophage Gal/GalNAc lectin 2 (MGL2)+ peritoneal antigen presenting cells during Fasciola hepatica infection are essential for regulatory T cell induction
Scientific Reports (2022)
-
Severity of neonatal influenza infection is driven by type I interferon and oxidative stress
Mucosal Immunology (2022)
-
STAT1-mediated induction of Ly6c-expressing macrophages are involved in the pathogenesis of an acute colitis model
Inflammation Research (2022)
-
The Effect of BTK Inhibitor Ibrutinib on Leishmania infantum Infection In Vitro
Acta Parasitologica (2022)
-
Leishmania donovani infection suppresses Allograft Inflammatory Factor-1 in monocytes and macrophages to inhibit inflammatory responses
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.