Abstract
The nuclear receptor Rev-erb-α modulates hepatic lipid and glucose metabolism, adipogenesis and thermogenesis. We have previously demonstrated that Rev-erb-α is also an important regulator of skeletal muscle mitochondrial biogenesis and function, and autophagy. As such, Rev-erb-α over-expression in skeletal muscle or its pharmacological activation improved mitochondrial respiration and enhanced exercise capacity. Here, in gain- and loss-of function studies, we show that Rev-erb-α also controls muscle mass. Rev-erb-α-deficiency in skeletal muscle leads to increased expression of the atrophy-related genes (atrogenes), associated with reduced muscle mass and decreased fiber size. By contrast, in vivo and in vitro Rev-erb-α over-expression results in reduced atrogenes expression and increased fiber size. Finally, Rev-erb-α pharmacological activation blocks dexamethasone-induced upregulation of atrogenes and muscle atrophy. This study identifies Rev-erb-α as a promising pharmacological target to preserve muscle mass.
Similar content being viewed by others
Introduction
Skeletal muscle is the most abundant tissue in the body, accounting for 30 to 40% of total body mass. Besides its role in locomotion and posture, skeletal muscle controls whole body energy metabolism by ensuring blood glucose control and is also the major source of amino acids (AAs) that can be used by various tissues during prolonged periods of fasting. Skeletal muscle is highly plastic and muscle mass, fiber type and size can undergo major remodeling to meet developmental and environmental challenges1. Muscle mass increases during development and in the adult in response to mechanical overload or hormonal stimulation, whereas muscle wasting is a common condition in diverse pathologies including obesity/type 2 diabetes, cancers, myopathies, chronic inflammatory disorders and is also a hallmark of aging, undernutrition or inactivity2,3. Chronic glucocorticoid therapies for the treatment of inflammatory or auto-immune disorders are also characterized by a rapid muscle atrophy. Loss of muscle mass results in reduced ability to sustain contraction, weakness of peripheral and respiratory muscles that compromises mobility and negatively affects quality of life. Muscle wasting also results in poor prognosis in diverse diseases, and ultimately has major clinical implications3. Therefore, understanding the signaling pathways that control muscle mass is of special interest for the development of novel therapies to preserve muscle mass.
Skeletal muscle mass reflects the delicate balance between protein synthesis and degradation. Protein synthesis is essentially controlled by the insulin-like growth factor 1– phosphoinositide-3-kinase–Akt/protein kinase B–mammalian target of rapamycin (IGF1-PI3K-Akt-mTOR) signaling pathway in response to a variety of stimuli such as IGF1, insulin, AAs derived from protein degradation, β2-adrenergic receptor agonists or androgens1. Activation of PI3K induces Akt phosphorylation which, in turn, inhibits the protein complex TSC1 (Tuberous Sclerosis Complex 1)-TSC2 thus activating the mTOR pathway through the two main downstream effectors of mTORC1: S6K (p70S6 kinase) and 4E-BP1 (eIF-4E-binding protein)4. pAkt also negatively regulates protein degradation by phosphorylating the Forkhead box O (FoxO) transcription factors and by promoting their exclusion from the nucleus1.
Protein degradation occurs via two proteolytic pathways that are tightly regulated by signaling pathways and transcription factors: the ubiquitin-proteasomal system, which predominantly breakdowns myofibrillar proteins, and the autophagic-lysosomal machinery, which ensures the maintenance of cellular homeostasis and the physiological turnover of damaged/aged cellular organelles1,3. Upon atrophy signals, FoxOs induce the transcription of the two E3 ubiquitin ligases MuRF1 (Muscle-Specific RING Finger Protein 1) and Atrogin-1 (MAFbx) to promote proteasomal protein degradation5,6. Mouse models invalidated for atrogin-1 or Murf1 are resistant to starvation and glucocorticoid (GC)-induced atrophy, respectively7,8,9,10. The FoxO family is composed of three isoforms FoxO1, FoxO3a and FoxO4 and in vivo upregulation of FoxO1 and FoxO3 is sufficient to promote muscle atrophy6,11,12,13. In addition, FoxO activation also induces autophagy14,11, a process tightly regulated at the gene, protein and flux levels and involving several autophagy-related genes (atg) as well as proteins such as Parkin and Bnip3 which contribute to mitochondrial remodeling15.
Circulating levels of GC are increased in diseases associated with muscle atrophy like sepsis or cachexia, and promote muscle mass loss16,17. Similarly, administration of synthetic GCs, such as dexamethasone used for the treatment of chronic inflammatory and immune diseases, results in muscle atrophy18. GCs promote both protein proteasomal degradation and autophagy-mediated proteolysis7,8,9. In addition, GCs exert anti-anabolic actions by up-regulating Regulated in Development and DNA damage responses 1 (Redd1, also known as ddit4) and Kruppel Like Factor 15 (KLF15), which directly repress mTOR signalling9,19,20. Beside, KLF15 acts on mTOR activity through the up-regulation of Branched Chain Amino Acid Transaminase 2 (BCAT2), a mitochondrial enzyme that degrades branched chain AAs. KLF15 also enhances Murf1 and atrogin-1 gene expression9.
The nuclear receptor Rev-erb-α acts as a transcriptional repressor by recruiting co-repressors and binding to specific DNA regions of its target genes21,22 or by recruiting co-repressors to sites to which it is tethered by tissue-specific transcription factors23. We and others have shed light on the role of Rev-erb-α in the circadian control of hepatic lipid, bile acid and glucose metabolism21,22,24,25,26,27, white adipocyte in vitro differentiation28,29, as well as brown adipocyte thermogenesis30. We have recently shown that Rev-erb-α controls skeletal muscle autophagy and mitochondrial biogenesis. Moreover, pharmacological activation of Rev-erb-α in vivo increases skeletal muscle mitochondrial respiration and exercise capacity, identifying Rev-erb-α as a target to improve muscle function31. Whether Rev-erb-α controls muscle mass has not been investigated yet.
Here, we report that Rev-erbα −/− mice display significantly reduced skeletal muscle mass and decreased fiber cross section area. This is associated with an increased expression of atrophy-related genes in skeletal muscle from Rev-erbα −/− mice, while AAV-mediated skeletal muscle-specific-Rev-erb-α overexpression confers an opposite phenotype. This effect on gene expression and fiber size was recapitulated in in vitro models of Rev-erb-α gain- and loss-of-function in myogenic cells. In addition, Rev-erb-α over-expression and/or pharmacological activation partially or totally blunted the induction of atrophy genes by dexamethasone in a mouse myoblast cell line as well as muscle mass loss in vivo. Together these data point to a new role for Rev-erb-α in the control of skeletal muscle mass and suggest that pharmacological activation of Rev-erb-α may constitute a promising approach to limit muscle wasting and improve the quality of life and survival of patients.
Results
Rev-erbα −/− mice display reduced muscle mass associated with smaller muscle fibers
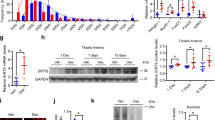
Here, we explored whether Rev-erb-α plays a role in the control of skeletal muscle mass or fiber size. Different hindlimb skeletal muscles were harvested from the Rev-erbα −/− mice and their Rev-erbα +/+ littermates and weighed. Quadriceps, gastrocnemius and tibialis anterior (TA) mass was significantly lower (by −20.9%, −14.5% and −11.5%, respectively) in Rev-erbα −/− compared to Rev-erbα +/+ mice (Fig. 1A), whereas body weights were similar (Figure S1). The weight of the soleus muscle, which mostly contains oxidative fibers, was also reduced in Rev-erbα −/− mice compared to Rev-erbα +/+ mice (by 12.6%) although the difference did not reach statistical significance (Fig. 1A). This is in accordance with the fact that glycolytic muscles are generally more vulnerable to atrophy than oxidative muscles19. Interestingly, Rev-erbα +/− mice which have normal locomotor activity also display a tendency to reduced muscle mass compared to the wild-type mice (data not shown), thus indicating that reduced muscle mass occurs independently of any changes in the locomotor activity.
Rev-erb-α controls skeletal muscle mass. (A) Quadriceps, gastrocnemius, tibialis anterior and soleus muscle mass from Rev-erbα +/+ and Rev-erbα −/− mice (n = 13 and 11, respectively). (B) Representative immunostaining of laminin on skeletal muscle sections (left) and mean fiber cross-sectional area and fiber area distribution of tibialis anterior muscle from Rev-erbα +/+ and Rev-erbα −/− mice (>13 000 fibers per mouse, n = 4 mice per genotype) (right). (C) Representative immunostaining of myosin Heavy Chain (left) and mean fiber diameter and fiber diameter distribution of fibers differentiated from myogenic precursors isolated from Rev-erbα +/+ and Rev-erbα −/− mice (3 measures per fiber, n = 120 fibers per genotype) (right). (D) Representative immunostaining of Myosin Heavy Chain (left) and mean fiber diameter and fiber diameter distribution of differentiated C2C12 cells over-expressing Rev-erbα (pREV-ERB-α) and control cells (pBabe) (3 measures per fiber, n = 120 fibers per group) (right). Results are expressed as means ± sem; *p < 0.05, **p < 0.01 by t-test (A), Mann-Whitney test (B–D) or chi-square test (B–D) to compare, respectively, the mean or the frequency distribution between groups.
To further characterize the impact of Rev-erb-α on skeletal muscle mass, skeletal muscle fiber cross sectional areas (CSA) were quantified on TA sections using a laminin staining which surrounds muscle fibers. In line with the reduction of muscle mass, the mean fiber CSA was significantly lower in Rev-erbα −/− compared to Rev-erbα +/+ mice with a shift in the fiber size distribution toward smaller fibers (Fig. 1B). These data therefore indicate that skeletal muscle is atrophied in absence of Rev-erb-α.
Next, primary myofibers were differentiated in vitro from myogenic precursors isolated from Rev-erbα +/+ and Rev-erbα −/− mice to assess the role of Rev-erb-α exclusively in myogenic cells. As observed in vivo, the mean muscle fiber diameter was significantly lower in Rev-erbα-deficient cells, again with a shift toward smaller fibers (Fig. 1C). Next, fiber diameter was measured on in vitro differentiated C2C12 overexpressing Rev-erb-α (pREV-ERB–α) and control C2C12 myofibers (pBabe). Expectedly, a mirror phenotype was observed, ie the mean fiber diameter was significantly higher in Rev-erb-α overexpressing fibers compared to controls, with a decrease in smaller fiber size and an increase in larger fibers (Fig. 1D). Thus, muscle mass and fiber size changes reflect a direct, cell autonomous, impact of Rev-erb-α on fibers rather than an adaptive response to a chronic situation.
Rev-erb-α negatively regulates catabolic and anti-anabolic pathways in skeletal muscle
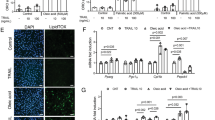
To determine whether these changes in muscle mass and fiber size are due to a dysregulation of the transcriptional programs elevated in muscle atrophy, expression analysis of the genes belonging to these processes was performed on skeletal muscles isolated from both Rev-erb–α loss- and gain-of-function mouse models. In particular, expression of genes encoding proteins involved in the ubiquitin-proteasome system was measured. Our results show that expression of the catabolic genes ubiquitin (Ubc), encoding the polyubiquitin precursor, and the two E3 ubiquitin ligases Atrogin and Murf1, as well as their transcriptional regulators Foxo1 and Foxo3a is significantly increased in muscle from Rev-erbα −/− mice compared to their Rev-erbα +/+ littermates (Fig. 2A). By contrast, AAV-mediated Rev-erb-α over-expression in wild-type mice (Figure S2A) led to the mirror phenotype, namely a significant downregulation of these catabolic genes (Fig. 2B, Figure S2). These results are in accordance with the increase in proteosomal activity in myofibers obtained after differentiation of myogenic precursor isolated from Rev-erbα +/+ and Rev-erbα −/− mice and the mirrored decreased proteasomal activity measured in C2C12 cells overexpressing Rev-erb-α (Figure S3).
Rev-erb-α controls atrophy-related genes in skeletal muscle. (A) RT-qPCR analysis of atrophy-related gene expression in quadriceps muscle from Rev-erbα +/+ and Rev-erbα −/− mice (n = 5 per genotype). (B) RT-qPCR analysis of atrophy-related gene expression in gastrocnemius muscle from mice intra-muscularly injected with a Rev-erb-α expressing or a control AAV vector (n = 7 per group). Results are expressed as means ± sem; *P < 0.05, **P < 0.01, ***P < 0.001 by Mann-Whitney test.
We also examined the expression of Redd1 and Klf15, two genes encoding negative regulators of the anabolic mTOR signaling pathway and known glucocorticoid receptor (GR) targets9. Expression of both genes was upregulated in skeletal muscle in absence of Rev-erbα (Fig. 2A), while Rev-erb-α over-expression decreased expression of these genes (Fig. 2B, Figure S2B). Accordingly, expression of Bcat2, a downstream Klf15 target gene, was significantly upregulated in Rev-erbα −/− skeletal muscle, and down-regulated upon Rev-erb-α over-expression (Fig. 2A,B, Figure S2B). These alterations occurred in absence of any changes in the mTOR signaling pathway assessed by western blot (data not shown).
Altogether with our previous observation that Rev-erb-α down-regulates skeletal muscle autophagy31, these findings indicate that Rev-erb-α represses catabolic and anti-anabolic pathways in skeletal muscle, and that its absence leads to skeletal muscle atrophy through the activation of these pathways.
Rev-erb-α over-expression and pharmacological activation counteracts atrophy induced by dexamethasone
Chronic dexamethasone treatment induces skeletal muscle fibers atrophy through the activation of different genes found to be regulated by Rev-erb-α. Therefore, we tested whether Rev-erb-α can counteract dexamethasone-induced muscle wasting. To this aim, Rev-erb-α over-expressing and control C2C12 myogenic cells were differentiated and then treated during 48 hrs with dexamethasone. Rev-erb-α overexpression partially or totally blunted dexamethasone-mediated induction of the catabolic (Atrogin, Murf1, Foxo1 and Foxo3a) and anti-anabolic (Klf15, Redd1 and Bcat2) genes (Fig. 3). Rev-erb-α is a ligand-activated nuclear receptor, and we have previously demonstrated that its pharmacological activation with the SR9009 compound has beneficial effects on skeletal muscle endurance training31. Therefore, we next assessed whether Rev-erb-α activation may preserve muscle mass upon glucocorticoid administration in vivo. To this aim, wild-type mice were treated with dexamethasone alone or with the Rev-erb agonist SR9009 for 3 days. As expected, dexamethasone treatment potently induced the expression of the catabolic genes (Atrogin, Murf1, Ubc, Foxo1 and Foxo3a) as well as the well-known Klf15, Redd 1 and Bcat2 GR target genes (Fig. 4A). Interestingly, co-treatment of the mice with the SR9009 Rev-erb-α ligand partially or totally blunted the induction of these genes by dexamethasone (Fig. 4A), resulting in muscle mass preservation (Fig. 4B). Quantification of muscle fiber diameter was performed upon 3 days treatment with dexamethasone on fully differentiated cells. Fiber diameter was strongly reduced upon dexamethasone treatment, and SR9009 co-treatment fully prevented this effect (Fig. 4C). Altogether, these data indicate that Rev-erb-α overexpression or its pharmacological activation abolishes dexamethasone atrophic effects in vitro and in vivo.
Rev-erb-α over-expression counteracts dexamethasone-mediated induction of atrophy-related genes. RT-qPCR analysis of atrophy-related gene expression in differentiated C2C12 cells infected with Rev-erb-α or control retrovirus and treated with dexamethasone (1 µM) or vehicle for 48hrs (n = 6 per condition). Results are expressed as means ± sem; **P < 0.01, ***P < 0.001 by One-way ANOVA with Bonferroni post-hoc analysis.
Pharmacological Rev-erb-α activation preserves muscle mass upon dexamethasone treatment. (A) RT-qPCR analysis of atrophy-related gene expression in quadriceps muscle (n = 5 per group) and (B) quadriceps muscle mass (n = 4 per group) from mice receiving or not 10 mg/kg/day of dexamethasone and co-treated with SR9009 (100 mpk) twice daily or vehicle for 3 days. (C) Mean fiber diameter and fiber size distribution of differentiated C2C12 cells treated or not with dexamethasone (20 µM) and co-treated with SR9009 (10 µM) or vehicle for 3 days (3 measures per fiber, n = 120 fibers per group). Results are expressed as means ± sem; *p < 0.05, **p < 0.01, ***p < 0.001 by One-way ANOVA with Bonferroni post-hoc analysis or chi-square test to compare respectively the mean or the frequency distribution between groups.
Rev-erb-α controls FoxOs nuclear translocation and directly targets atrophy-related genes
Dexamethasone treatment promotes nuclear translocation of FoxO1 and FoxO3a. Once in the nucleus, they activate the expression of their target genes such as Atrogin or Murf1. To determine if pharmacological activation of Rev-erb-α with the SR9009 is able to modulate FoxOs cellular localization, C2C12 cells were treated with dexamethasone and co-treated or not with SR9009 and the intensity of FoxO1 and FoxO3a signal was analysed by immunofluorescence and quantified. Results show that the nuclear intensity of both FoxO1 and FoxO3a is significantly increased upon dexamethasone treatment, whereas SR9009 co-treatment blunted dexamethasone-induced FoxO1 and FoxO3a nuclear translocation (Fig. 5A,B).
Rev-erb-α modulates FoxO nuclear translocation and directly regulates atrophy-related genes through a direct binding to their regulatory sequences. (A) Representative immunostaining of FoxO1 (upper panels) and FoxO3a (lower panels) in green and Dapi nuclear staining in blue of C2C12 myogenic cells treated or not with dexamethasone (1 µM) and co-treated with SR9009 (10 µM). (B) Nuclear intensity quantification of FoxO1 and FoxO3a (n > 100 cells per condition). Results are expressed as means ± s.e.m; *p < 0.05, ***p < 0.001, ****p<0.0001 by One-way ANOVA with Bonferroni post-hoc analysis. (C) Rev-erb-α binding to regulatory regions of the indicated atrophy-related genes measured by ChIP-qPCR (n ≥ 2 independent experiments). Data are expressed as means ± s.e.m.
Beside its action through FoxO signaling, Rev-erb-α may directly repress its target genes by binding specific DNA sequences in their regulatory regions. Results from ChIP experiments performed on C2C12 myogenic cells show that Rev-erb-α occupies a site located at −2 kb from the transcription star site (TSS) within the promoter sequence of Bcat2 gene and a site located at −2.5 kb from the TSS in Murf1 gene regulatory sequences (Fig. 5C).
In conclusion, these data show that Rev-erb-α counteracts atrophy through the modulation of FoxOs translocation, thus preventing their transcriptional activation and by a direct binding to atrophy-related genes.
Discussion
Skeletal muscle serves several important functions in the body, from locomotion and posture to metabolic homeostasis. Muscle loss is observed in and aggravates diverse physiological (such as fasting or aging) and pathological situations (such as cancer, sepsis, inflammatory conditions), and is associated with a poor quality of life and prognosis3. Thus, preservation of muscle mass may have major clinical benefits.
In this study, we addressed the role of the drugable nuclear receptor Rev-erb-α in the control of muscle mass. We demonstrated that Rev-erb-α modulates muscle mass and fiber size in vivo and in vitro by repressing the expression of genes of the ubiquitin-proteasomal degradation pathway such as Ubc, the atrogin-1 and MuRF1E3 ubiquitin ligases, as well as the upstream FoxO1 and FoxO3a regulators. Our data support an intrinsic role for Rev-erb-α in the control of muscle mass. Indeed, in vitro observation that muscle fibers differentiated from Rev-erbα −/− primary myogenic progenitors display similar alterations (decreased fiber size and upregulated expression of atrophy genes), whereas Rev-erb-α over-expression in C2C12 myoblasts led to the opposite phenotype, demonstrate a cell autonomous action of Rev-erb-α in regulating muscle atrophy.
Atrogin-1 and MuRF1 mediate myofibrillar protein degradation, and nutrient-deprivation–induced and dexamethasone-induced muscle atrophy, respectively5,6,7,10,32. These two genes are regulated by FoxO transcription factors whose upregulation in vivo promotes muscle atrophy6,12,13. Indeed, the expression of a constitutively active FoxO3 mutant up-regulates Atrogin-1 transcription and induces skeletal muscle atrophy, whereas a dominant negative FoxO3 prevents fasting-induced Atrogin-1 expression and dexamethasone-induced atrophy in myotubes6. In line, FoxO1,3,4 deletion prevented muscle loss and weakness upon fasting11. In our study, we show that Rev-erb-α represses these genes and controls FoxO nuclear translocation, thereby controlling muscle mass. FoxOs also control the lysosome-autophagy pathway that, beside the ubiquitin-proteasomal system, plays a critical role in skeletal muscle atrophy33. FoxOs mediate activation of some autophagy genes and proteins such as Bnip3, LC-3, and cathepsin L, and fasting failed to induce autophagosome formation in FoxO-deficient mice11. Interestingly, we have already demonstrated that Rev-erbα-deficiency in skeletal muscle increased expression of several autophagy genes such as Bnip3, Pink and Parkin, and increased autophagy flux as shown by increased LC3-II/LC3-I ratio31. This was also observed in Rev-erbα-deficient myoblasts, whereas Rev-erb-α over-expression led to a strong repression of autophagy genes. Besides being recruited to the promoter of several autophagy genes (Bnip3 and Cathepsin L, among others)31, we show here that Rev-erb-α may also regulate this pathway in an indirect manner through the repression of FoxOs. Thus, our previous study and the present report demonstrate that Rev-erb-α plays a prominent role in the atrophy program by regulating the ubiquitin-proteasome and autophagy pathways in a coordinated manner.
Other transcription regulators have been shown to modulate muscle mass, amongst which the cofactor PGC1-α. PGC1α expression is decreased in muscle atrophy and its over-expression inhibits FoxO3 signaling, autophagy and the ubiquitin-proteasome degradation pathways without affecting protein synthesis34. In addition, the expression of gene encoding oxidative phosphorylation is reduced in atrophying muscle suggesting that mitochondrial dysfunction can negatively impact skeletal muscle mass maintenance by triggering catabolic pathways35. It is noteworthy that Rev-erb-α positively controls PGC1α expression and activity, as well as mitochondrial biogenesis and function in skeletal muscle31, which may contribute to the changes in muscle mass observed in the Rev-erb-α gain- and loss-of function models.
Endogenous GCs induce a moderate and reversible skeletal muscle protein catabolism in response to infection, whereas chronic GC treatment elicits muscle atrophy by increasing pathways leading to protein degradation and by antagonizing the action of anabolic factors in an exacerbated manner. In skeletal muscle, GR inhibits PI3K/Akt signaling, resulting in increased FoxOs signaling6,36. In addition, GR antagonizes the anabolic pathway by inducing Redd1 expression, which leads to mTORC1 inhibition37, and Klf15, which increases the expression of the branched AAs-degrading enzyme BCAT2, thereby strengthening the inhibition of mTOR activity in an Akt-independent manner9,38. KLF15 also represses Murf1 and Atrogin-1 gene expression either directly for Murf1 or through FoxO induction for Atrogin-1 9. Corticosterone levels are not different in Rev-erbα-deficient mice39,40. However, Rev-erbα gene expression is down-regulated in skeletal muscle upon dexamethasone treatment (Figure S4). We and others have already evidenced a regulation of Rev-erbα by GR and the cross-talk between these two nuclear receptors in the liver40,41, and it may be hypothesized that Rev-erbα down-regulation, or gene ablation, is permissive to GR action on atrophy genes. This is in line with the decreased muscle mass in the Rev-erbα-deficient mice in basal conditions and with the repression of FoxOs (and both proteasomal and autophagy-related genes), and of Redd1, Klf15 and Bcat2 by Rev-erb-α. In agreement with this concept, we demonstrate that Rev-erb-α pharmacological activation is able to counteract dexamethasone-mediated induction of these genes, thereby preserving muscle mass. Our study identifies Rev-erb-α as a new actor in the complex network that controls muscle atrophy and a promising target to preserve muscle mass beside its beneficial effect on exercise capacity.
Methods
Animals
7–12 week-old Rev-erbα-deficient mice (Rev-erbα −/−) and their wild-type (Rev-erbα +/+) littermates were studied. Mice were euthanized by cervical dislocation. Skeletal muscles (Gastrocnemius, Quadriceps, Tibialis Anterior (TA) and Soleus) were weighed and snap-frozen.
To induce skeletal muscle atrophy, 7–8 weeks old wild-type mice received an intraperitoneal injection of dexamethasone at 10 mg/kg/day or vehicle (ethanol) for 3 consecutive days at 9:00 am. Each day, half of the dexamethasone-treated mice also received two injections of the SR9009 Rev-erb agonist at 100 mg/kg at 17:30 pm the previous day and 30 min prior to dexamethasone injection. Quadriceps muscles were collected and weighed.
To induce Rev-erb-α overexpression (Figure S2A), the Nr1d1 coding sequence was introduced in an AAV1 vector (Penn Vector Core, University of Pennsylvania) and 3 × 1011 genome copies were injected intra-muscularly in the TA and gastrocnemius muscles of wild-type mice as previously described31. The contralateral limb was injected with control AAV. 6 weeks later, mice were sacrificed and muscles were snap-frozen and stored until mRNA extraction.
Mice were housed in a 12 h/12 h light/dark cycle and had free access to food and water. All experimental procedures were performed with the approval of the Nord-Pas-de-Calais ethics committee (CEEA75) and in compliance with French and European ethical legislations.
Myogenic Cell Culture
TA, gastrocnemius, soleus, quadriceps, diaphragm, triceps and pectoralis muscles were collected from 7–8 weeks- old Rev-erbα-deficient mice (Rev-erbα −/−) and their wild-type littermates (Rev-erbα +/+) and digested in Dulbecco’s modified Eagle’s medium (DMEM; 4.5 g/L D Glucose; Gibco) F12 medium with Collagenase D (0.15 U/mg) and Dispase II (0.8 U/mg) during 30 minutes at 37 °C. Digested muscles were then filtered on cell strainer and myogenic precursors were isolated using negative and positive selection kits (130104268 and 130104261, Miltenyi Biotec). Primary myoblasts were resuspended in DMEM F-12 and with 20% FBS, 1% gentamycin, 1 mL Ultroser G and 5 ng/mL FGFb and seeded in 0.1% gelatin coated plates. After 3 days, differentiation was induced by the addition of F-12/DMEM (1:1) medium supplemented with 5% horse serum for 7 days.
Constitutively over-expressing Rev-erb-α (REV-ERB-α) and control (pBabe) C2C12 cell lines31 were cultured in DMEM medium with 1% gentamycin and 10% FBS. At 80% of confluency, myogenic differentiation was induced by addition of 2% horse serum for 3 days.
For myotube atrophy experiments, C2C12 cells were cultured as previously described31. After 5 days of differentiation, cells were treated with dexamethasone (20 µM)42 or vehicle (ethanol), and SR9009 (10 µM) as indicated for 3 days. For analysis of atrophy-related gene expression, C2C12 were treated with dexamethasone (1 µM) during 2 days with or without SR9009 (10 µM). For FoxOs nuclear translocation analysis, C2C12 were treated with dexamethasone 1 µM or vehicle with or without SR9009 (10 µM) overnight.
Immunostaining
C2C12 cells were fixed with 4% paraformaldehyde, and permeabilized with 0.2% Triton × 100/ 50 mM NH4Cl in PBS. Cells were then incubated with Myosin Heavy Chain (MF-20) (1:250, DSHB AB2147781), FoxO1 (1:250, Santa Cruz 2880 S) or FoxOa3 (1:250, Santa Cruz 2497S) antibodies, followed by Alexa Fluor 488-labelled secondary antibody (ThermoFisher Scientific A-11018) and Dapi. Cells were mounted with Dako Fluorescent Mounting Medium (S3023). Images were captured with a LSM 880 confocal microscope (Zeiss).
Immunofluorescence staining was also performed on TA sections (12 µm) after fixation with 4% paraformaldehyde and blocking with 5% horse serum in PBS. Sections were incubated with a laminin (1:250, Santa Cruz SC59854) primary antibody followed by Alexa Fluor 488-labelled secondary antibody (ThermoFisher Scientific A21208). Sections were mounted with Dako Fluorescent mounting medium (Dako S3023). Images were captured with an Axioscan (Zeiss).
Immunostaining Analysis
The area of muscle fibers and the diameter of primary and C2C12 myofibers were quantified using a FiJi macro43 for image input and preprocessing functions. For the diameter of primary and C2C12 myofibers, between 60 and 160 fibers were analyzed for each condition, and 3 measures were taken manually for each fiber and averaged.
The Nuclear intensity of FoxO1 and FoxO3a staining was performed using AcapellaTM image analysis software (version 2.7-Perkin Elmer Technologies, Waltham, USA) by using dedicated module to detect nuclei shape in Dapi following by the intensity tracking by nucleus in channel FITC. A minimum of 100 cells per condition were analyzed in two independent experiments.
Gene Expression Analysis
Total RNA was extracted using the guanidium thiocyanate/phenol/chloroform extraction method for frozen muscles and using trizol (15596018, Life technologies) for C2C12 cells as previously described31. DNAse treatment was performed (EN0521, Thermo Scientific) and cDNA was obtained using a reverse transcription kit (High-capacity cDNA reverse transcription Kit, Life Technologies). Quantitative qPCR was performed using specific primers (Supplemental Table 1) and the Brillant II SYBR Green QPCR Master Mix (Agilent Technologies). Results were analyzed with the standard delta Cycle Threshold method and normalized to the expression of PPia (Cyclophilin A).
Chromatin ImmunoPrecipitation (ChIP)
ChIP experiments were performed as previously described44 using the anti-Rev-erb-α (1:50, #13418, Cell Signalling) and normal rabbit IgG (1:250, #2729, Cell Signalling) antibodies following manufacturer’s instructions. Briefly, differentiated C2C12 cells were crosslinked with 1% paraformaldehyde for 10 min. Extracted nuclei were resuspended in lysis buffer (Tris-HCl 50 mM pH8.0, EDTA 10 mM, SDS 1%, protease inhibitor cocktail). Chromatin was then sheared for 20 min using a water-cooled Bioruptor (Diagenode) and diluted 10 times with the dilution buffer (Tris HCl 20 mM pH8.0, Triton X-100 1%, Glycerol 5%, EDTA 2 mM, NaCl 150 mM, protease inhibitor cocktail from Roche). 100 µg of DNA were used per IP and recovered chromatin was purified using QIAquick PCR clean up kit (Qiagen). Immunoprecipitated DNA was analysed by qPCR and normalized to input samples and negative DNA region.
Proteasome activity
The 20S Proteasome Activity Assay kit (APT, Millipore, Billerica, MA, USA) was used to measure the proteasome activity according to the manufacturer’s instructions.
Statistics
To compare two groups, data were analysed using the Mann-Whitney test or the two-tailed unpaired student’s t-test after being assessed for normality. One-way ANOVA with Bonferroni post-hoc analysis was used to compare more than 2 groups. Qualitative traits (i.e. area frequency distribution) were analysed with a Chi2 test. Statistical significance is shown on the graphs (*p < 0.05; **p < 0.01; ***p < 0.001; ****p<0.0001). Statistical methodologies used for each data set are indicated in the figure legends. Prism 6.0 (GraphPad Software Inc., USA) was used for statistical analysis.
References
Schiaffino, S., Dyar, K. A., Ciciliot, S., Blaauw, B. & Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J 280, 4294–4314 (2013).
Bodine, S. C. & Furlow, J. D. Glucocorticoids and Skeletal Muscle. Adv. Exp. Med. Biol. 872, 145–176 (2015).
Cohen, S., Nathan, J. A. & Goldberg, A. L. Muscle wasting in disease: molecular mechanisms and promising therapies. NatRevDrug Discov 14, 58–74 (2015).
Bodine, S. C. et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. NatCell Biol 3, 1014–1019 (2001).
Bodine, S. C. et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 (2001).
Sandri, M. et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 (2004).
Baehr, L. M., Furlow, J. D. & Bodine, S. C. Muscle sparing in muscle RING finger 1 null mice: response to synthetic glucocorticoids. J.Physiol 589, 4759–4776 (2011).
Sandri, M. et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc.Natl.Acad.Sci.USA 103, 16260–16265 (2006).
Shimizu, N. et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab 13, 170–182 (2011).
Cong, H., Sun, L., Liu, C. & Tien, P. Inhibition of atrogin-1/MAFbx expression by adenovirus-delivered small hairpin RNAs attenuates muscle atrophy in fasting mice. HumGene Ther 22, 313–324 (2011).
Milan, G. et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 6, 6670 (2015).
Kamei, Y. et al. Skeletal muscle FOXO1 (FKHR) transgenic mice have less skeletal muscle mass, down-regulated Type I (slow twitch/red muscle) fiber genes, and impaired glycemic control. J. Biol. Chem. 279, 41114–41123 (2004).
Sandri, M. Regulation and involvement of the ubiquitin ligases in muscle atrophy. Free Radic. Biol. Med. 75(Suppl 1), S4 (2014).
Zhao, J. et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6, 472–483 (2007).
Mizushima, N. & Komatsu, M. Autophagy: renovation of cells and tissues. Cell 147, 728–741 (2011).
Braun, T. P. et al. Central nervous system inflammation induces muscle atrophy via activation of the hypothalamic-pituitary-adrenal axis. J. Exp. Med. 208, 2449–2463 (2011).
Lecker, S. H., Solomon, V., Mitch, W. E. & Goldberg, A. L. Muscle protein breakdown and the critical role of the ubiquitin-proteasome pathway in normal and disease states. J. Nutr. 129, 227S–237S (1999).
Braun, T. P. & Marks, D. L. The regulation of muscle mass by endogenous glucocorticoids. Front Physiol 6, 12 (2015).
Schakman, O., Kalista, S., Barbe, C., Loumaye, A. & Thissen, J. P. Glucocorticoid-induced skeletal muscle atrophy. IntJBiochemCell Biol 45, 2163–2172 (2013).
Britto, F. A. et al. REDD1 deletion prevents dexamethasone-induced skeletal muscle atrophy. AmJPhysiol EndocrinolMetab 307, E983–E993 (2014).
Bugge, A. et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev 26, 657–667 (2012).
Cho, H. et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature 485, 123–127 (2012).
Zhang, Y. et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 348, 1488–1492 (2015).
Duez, H. & Staels, B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab.Vasc.Dis.Res. 5, 82–88 (2008).
Duez, H. et al. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology 135, 689–698 (2008).
Raspe, E. et al. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. JLipid Res 43, 2172–2179 (2002).
Yin, L. et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789 (2007).
Fontaine, C. et al. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) gamma target gene and promotes PPARgamma-induced adipocyte differentiation. J.Biol.Chem. 278, 37672–37680 (2003).
Wang, J. & Lazar, M. A. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol 28, 2213–2220 (2008).
Gerhart-Hines, Z. et al. The nuclear receptor Rev-erbalpha controls circadian thermogenic plasticity. Nature 503, 410–413 (2013).
Woldt, E. et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat. Med. 19, 1039–1046 (2013).
Clarke, B. A. et al. The E3 Ligase MuRF1 degrades myosin heavy chain protein in dexamethasone-treated skeletal muscle. Cell Metab. 6, 376–385 (2007).
Mammucari, C. et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6, 458–471 (2007).
Brault, J. J., Jespersen, J. G. & Goldberg, A. L. Peroxisome proliferator-activated receptor gamma coactivator 1alpha or 1beta overexpression inhibits muscle protein degradation, induction of ubiquitin ligases, and disuse atrophy. J. Biol. Chem. 285, 19460–19471 (2010).
Romanello, V. & Sandri, M. Mitochondrial Quality Control and Muscle Mass Maintenance. Front. Physiol. 6, 422 (2015).
Stitt, T. N. et al. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14, 395–403 (2004).
Wang, H., Kubica, N., Ellisen, L. W., Jefferson, L. S. & Kimball, S. R. Dexamethasone represses signaling through the mammalian target of rapamycin in muscle cells by enhancing expression of REDD1. J. Biol. Chem. 281, 39128–39134 (2006).
Waddell, D. S. et al. The glucocorticoid receptor and FOXO1 synergistically activate the skeletal muscle atrophy-associated MuRF1 gene. Am. J. Physiol. Endocrinol. Metab. 295, E785–797 (2008).
Delezie, J. et al. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 26, 3321–3335 (2012).
Okabe, T. et al. REV-ERBα influences the stability and nuclear localization of the glucocorticoid receptor. J. Cell Sci. 129, 4143–4154 (2016).
Torra, I. P. et al. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology 141, 3799–3806 (2000).
Han, D.-S., Yang, W.-S. & Kao, T.-W. Dexamethasone Treatment at the Myoblast Stage Enhanced C2C12 Myocyte Differentiation. Int. J. Med. Sci. 14, 434–443 (2017).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Pourcet, B. et al. LXRα regulates macrophage arginase 1 through PU.1 and interferon regulatory factor 8. Circ Res. 109, 492–501 (2011).
Acknowledgements
We acknowledge the support from INSERM, Contrat Plan Etat Région (CPER), the Région Nord Pas-de-Calais/FEDER, the EU consortium Eurhythdia (FP7), the ANR-Labex-EGID (EGID, ANR-10-LABX-46), EFSD (European Foundation for the Study of Diabetes)-Eli Lilly, the Fondation Francophone pour la recherche sur le diabète (FFRD) together with the Fédération Française des Diabétiques (AFD), AstraZeneca, Eli Lilly, Merck Sharp & Dohme (MSD), Novo Nordisk & Sanofi, an 'ERC-Région Hauts de France' grant to H.D. and a grant from Fondation de France. A.M.L. was supported by the Association Française contre les Myopathies (AFM). We thank the BICeL facility for access to systems and technical advice.
Author information
Authors and Affiliations
Contributions
A.M.L., Q.T., S.D., J.B., C.D., A.D., S.L., B.P., E.W., A.B., L.P., M.Z., Y.S. and H.D. performed the experiments. A.D gave conceptual advice. A.M.L., Q.T., B.S., Y.S. and H.D. wrote the manuscript. A.M.L., Q.T., Y.S. and H.D. contributed to the study concept, research design and interpretation of the results. Y.S. and H.D. are the guarantors of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors contributed to discussion, reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mayeuf-Louchart, A., Thorel, Q., Delhaye, S. et al. Rev-erb-α regulates atrophy-related genes to control skeletal muscle mass. Sci Rep 7, 14383 (2017). https://doi.org/10.1038/s41598-017-14596-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-14596-2
This article is cited by
-
The role of REV-ERB in NASH
Acta Pharmacologica Sinica (2022)
-
Exercise alters the circadian rhythm of REV-ERB-α and downregulates autophagy-related genes in peripheral and central tissues
Scientific Reports (2022)
-
Rev-erbα exacerbates hepatic steatosis in alcoholic liver diseases through regulating autophagy
Cell & Bioscience (2021)
-
Transcriptome analysis of gravitational effects on mouse skeletal muscles under microgravity and artificial 1 g onboard environment
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.