Abstract
Kohlschutter-Tönz syndrome (KTS) is a rare genetic disorder with neurological dysfunctions including seizure and intellectual impairment. Mutations at the Rogdi locus have been linked to development of KTS, yet the underlying mechanisms remain elusive. Here we demonstrate that a Drosophila homolog of Rogdi acts as a novel sleep-promoting factor by supporting a specific subset of gamma-aminobutyric acid (GABA) transmission. Rogdi mutant flies displayed insomnia-like behaviors accompanied by sleep fragmentation and delay in sleep initiation. The sleep suppression phenotypes were rescued by sustaining GABAergic transmission primarily via metabotropic GABA receptors or by blocking wake-promoting dopaminergic pathways. Transgenic rescue further mapped GABAergic neurons as a cell-autonomous locus important for Rogdi-dependent sleep, implying metabotropic GABA transmission upstream of the dopaminergic inhibition of sleep. Consistently, an agonist specific to metabotropic but not ionotropic GABA receptors titrated the wake-promoting effects of dopaminergic neuron excitation. Taken together, these data provide the first genetic evidence that implicates Rogdi in sleep regulation via GABAergic control of dopaminergic signaling. Given the strong relevance of GABA to epilepsy, we propose that similar mechanisms might underlie the neural pathogenesis of Rogdi-associated KTS.
Similar content being viewed by others
Introduction
Neurological disorders caused by single-gene mutations are important genetic models to understand how individual genes execute their roles to support the development and function of the brain, as in the case of Kohlschutter-Tönz syndrome (KTS). KTS patients display developmental delays and psychomotor regression1. The most prominent symptoms include amelogenesis imperfecta, early-onset seizures, and intellectual disabilities. Linkage analyses followed by genomic sequencing have revealed that most, if not all, KTS patients have homozygous nonsense, frameshift deletion, or splicing site mutations at the Rogdi locus2,3,4,5,6. ROGDI protein expression is not detectable in affected individuals2, indicating that the loss of Rogdi function is responsible for the pathogenesis of KTS. In wild-type human tissues, Rogdi transcripts are ubiquitously expressed while the highest enrichment is evident in adult brain and spinal cord2, 3. This observation is consistent with the neurological phenotypes observed in KTS patients.
Given that Rogdi homologs are relatively well conserved in higher eukaryotes, animal models may facilitate our understanding of Rogdi-dependent neural processes. In fact, Rogdi was initially identified in a Drosophila genetic screen as a memory-relevant gene and was thus named after one of Pavlov’s dogs7. Sequence analyses of Rogdi homologs revealed a putative leucine zipper (ZIP) motif, which could mediate the dimerization of DNA-binding basic ZIP (bZIP) transcription factors5. Interestingly, ROGDI proteins localize to the nuclear envelope in cultured human cells2, although they lack basic amino acid residues that are typically located at the N-terminus of the ZIP domain and are required for the DNA-binding and nuclear localization of the bZIP transcription factors8. Nonetheless, few or no studies have demonstrated the biological activity of Rogdi 9 and genetic models for Rogdi homologs have not been reported yet. Therefore, how Rogdi exerts its physiological roles particularly in the central nervous system and how its mutation leads to the development of KTS are largely unknown.
In the course of our genetic studies to elucidate genes and regulatory pathways involved in sleep behaviors, we identified novel sleep mutant alleles in the Drosophila Rogdi gene. Here, we employed the sleep-promoting effects of Rogdi as a readout of its neural function and demonstrated that Rogdi acts cell-autonomously in GABAergic neurons to enhance metabotropic GABA transmission and thereby sustain sleep. In addition, dopaminergic rescue of Rogdi mutant sleep revealed a novel sleep-regulatory mechanism that functionally links a specific subset of sleep-promoting GABAergic neurons to a wake-promoting dopaminergic pathway. Since epilepsy, a well penetrated phenotype in KTS patients, implicates GABAergic transmission10,11,12 and sleep disorders13,14,15,16, our findings provide an important genetic clue to understanding the molecular and neural pathogenesis of KTS.
Results
Loss-of-function Mutations in Rogdi Suppress Sleep Behaviors in Drosophila
Sleep is physiologically essential for animals. Genetic models for sleep and sleep-related disorders have been established in multiple species in order to understand why we sleep and how sleep homeostasis occurs17,18,19. In addition, sleep-relevant genes have been successfully established in Drosophila sleep models by isolating novel sleep mutants and characterizing their sleep-modulatory effects20,21,22,23,24,25. We took a similar approach to identify a transgenic fly that harbors a genomic insertion of a transposable P element and displays insomnia-like sleep behaviors in 12-hour light: 12-hour dark (LD) cycles (Fig. 1). Sequence mapping of the transgene revealed insertion of the P element in the third intron at the Rogdi locus (Rogdi P1) (Fig. 1a). We further excised the Rogdi P1 insertion to generate a ~1 kb genomic deletion allele (Rogdi del) that caused an in-frame deletion of amino acid residues 16–83 in the gene product. Mutant flies homozygous for either the Rogdi insertion or the deletion allele barely expressed ROGDI proteins in head extracts as assessed by immunoblotting with polyclonal anti-ROGDI antibody (Fig. 1b, Supplementary Fig. 1). To determine if the in-frame deletion caused by Rogdi del allele could give rise to mutant ROGDI proteins, we overexpressed the cDNAs corresponding to either wild-type or Rogdi del allele in Drosophila S2 cells and examined their protein products using an epitope-tag. Indeed, wild-type and ROGDIdel proteins were comparably detectable in immunoblots of total cell extracts but the mutant ROGDIdel proteins had much shorter half-life (~2.6 hours) than wild-type (~49.5 hours) (Fig. 1c, Supplementary Fig. 2). Moreover, ROGDIdel proteins were localized to distinct, cytoplasmic inclusions (Supplementary Fig. 3a). This subcellular localization contrasted with that of wild-type ROGDI proteins which were evenly distributed in both nucleus and cytoplasm of S2 cells or adult fly neurons (Supplementary Fig. 3b). Collectively, these data indicate that the Rogdi del allele substantially affects the proteostasis of ROGDI proteins in general (See Discussion).
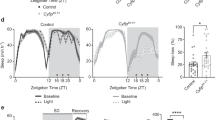
Rogdi mutation suppresses sleep in Drosophila. (a) A schematic of Rogdi mutant alleles. P element insertion (Rogdi P1) and genomic deletion (Rogdi del) are depicted in the context of the Rogdi locus. (b) Fly head extracts from wild-type and Rogdi mutants were immunoblotted with anti-ROGDI (top) and anti-TUBULIN (bottom, loading control) antibodies. Images were cropped from full-length blots shown in Supplementary Fig. 1. Protein size markers were shown on the right. (c) Wild-type ROGDI or ROGDIdel proteins with a 3xFLAG epitope tag were transiently expressed in Drosophila S2 cells and their decay rate (i.e., half-life) was measured after cycloheximide (CHX)-induced blockade of protein synthesis. Data represent average ± SEM (n = 3). Two-way ANOVA detected significant effects of genotypes (F[1,24] = 82.58, P < 0.0001). **P < 0.01, ***P < 0.001 to wild-type at each time-point as determined by Bonferroni post hoc test. (d) Daily sleep profiles of control and Rogdi mutant flies in 30 minute intervals. Sleep behaviors in individual flies were quantitatively analyzed under 12-hour light (white bar): 12-hour dark (black bar) (LD) cycles at 25 °C. Data represent average ± SEM (n = 61–126). ZT, zeitgeber time (lights-on at ZT0; lights-off at ZT12). (e) Daily sleep amount, average sleep bout length (ABL), and sleep latency in homozygous and trans-heterozygous Rogdi mutants were compared with those in wild-type and heterozygous controls. ***P < 0.001 to wild-type and heterozygous controls as determined by one-way ANOVA, Tukey post hoc test.
Importantly, the daily amount of sleep was substantially decreased in flies homozygous or trans-heterozygous for the mutant Rogdi alleles (Fig. 1d,e). The two mutant alleles led to distinct sleep profiles, likely due to the difference in their allelic nature. The sleep suppression phenotype was accompanied by sleep fragmentation: the average sleep bout length (ABL) was shorter in Rogdi mutants than in control flies (Fig. 1e). In addition, Rogdi mutants took longer to initiate their first sleep bout after lights off when looking at the sleep latency. Sleep fragmentation was also evident in female flies homozygous for the Rogdi deletion (Supplementary Fig. 4). However, Rogdi mutant phenotypes regarding total sleep amount and sleep latency were observed in virgins but not in mated females, possibly indicating that reproduction and/or neural processes relevant to post-mating responses might modulate Rogdi effects on sleep in female flies26, 27. Nonetheless, these data suggest that Rogdi is a novel sleep-promoting gene important for both sleep initiation and maintenance.
Insomnia-like Behaviors in Rogdi Mutants Do Not Implicate Deficits in Circadian Rhythms or Sleep Homeostasis
Circadian rhythms and sleep homeostasis have long been considered as two essential processes that shape sleep behaviors28. To understand how these two factors are implicated in Rogdi-dependent sleep, we examined Rogdi mutant sleep in different environmental or genetic conditions. The sleep suppression by Rogdi mutation persisted either in constant darkness or in a genetic background that harbored a hemizygous loss-of-function mutant allele of the circadian clock gene period 29, 30 (Fig. 2). Since the clock-less per mutants do not display daily oscillations in clock gene expression and locomotor behaviors, these data imply that Rogdi effects on sleep require neither LD cycles nor the functionality of circadian rhythms.
Wake-promoting effects of Rogdi mutation require neither LD cycles nor the functionality of circadian rhythms. (a,b) Daily sleep profiles of control and Rogdi mutant flies in wild-type (a) or clock-less per 01 mutant backgrounds (b). Sleep behaviors in individual flies were monitored in LD cycles followed by constant dark (DD) cycles at 25 °C. Data represent average ± SEM (n = 35–126). (c,d) Daily sleep amount (top) and average sleep bout length (ABL, bottom) in LD (c) or the first DD (d) cycles were compared among control and Rogdi mutant flies in wild-type or per 01 mutant backgrounds. ***P < 0.001 to control or per 01 mutants as determined by one-way ANOVA, Tukey post hoc test.
To examine whether Rogdi contributes to sleep homeostasis, we employed a mechanical sleep-deprivation protocol31. We optimized the visibility of homeostatic sleep regulation by applying 9 hours (rather than overnight) of mechanical stimulus starting from lights-off in the LD cycle. Sleep rebound was then measured for the last 3 hours in the D phase so that lights-on would not mask the compensatory increase in sleep drive after sleep deprivation. Under these conditions, Rogdi mutants displayed higher sleep rebound than control flies as assessed by the percentage of sleep gain during the recovery period (Fig. 3). While we do not exclude the possibility that longer baseline sleep in control flies and possible ceiling effects could have underestimated their relative sleep rebound, these data imply that Rogdi suppresses homeostatic sleep gain after sleep loss. In fact, this phenotype contrasts with sleep-promoting effects of Rogdi on baseline sleep but is consistent with a previous observation that sleep-regulatory pathways for baseline and recovery sleep could be genetically separable (See Discussion)32.
Rogdi mutation does not dampen sleep rebound after mechanical sleep deprivation. (a) Sleep behaviors in individual flies were monitored in LD cycles at 25 °C. Where indicated, flies were subject to the mechanical stimulus for the first 9 hours in the D phase (from ZT12 to ZT21) and their sleep rebound was quantitatively analyzed for the last 3 hours in the D phase (from ZT21 to ZT24). Averaged sleep profiles on the day of mechanical sleep deprivation (SD) (darker lines) were overlaid by those on the previous day (lighter lines). ZT, zeitgeber time (lights-on at ZT0; lights-off at ZT12). (b) Cumulative sleep loss was calculated in individual flies and averaged per each genotype. To calculate %sleep loss at each time-point, total sleep amount from ZT12 to a given time-point on the day of SD was subtracted by that on the previous day and then normalized to the total sleep amount from ZT12 and ZT21 on the previous day. Data represent average ± SEM (n = 31–40). Two-way repeated-measures ANOVA detected significant effects of genotypes (F[2,100] = 4.707, P = 0.0111) and their interaction with time-points (F[46,2300] = 2.495, P < 0.0001). **P < 0.01, ***P < 0.001 to control at each time-point as determined by Dunnett post hoc test.
A Wake-promoting Dopaminergic Pathway Mediates Rogdi Effects on Sleep
To elucidate the neural basis underlying the wake-promoting effects of Rogdi mutation, we first examined whether Rogdi mutants could be rescued by pharmacological treatment targeting specific neurotransmitter pathways. Tyrosine hydroxylase (TH) is a rate-limiting enzyme for the conversion of L-tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA), a precursor of the neurotransmitter, dopamine (DA) (Fig. 4a). Dopaminergic signaling pathways have been well established as having wake-promoting effects in both Drosophila and mammals21, 33, 34. In particular, a subset of TH-expressing dopaminergic neurons and their downstream postsynaptic target (D1-like DA receptor-expressing cells in the dorsal fan-shaped body) constitute a neural circuit that is important for promoting wakefulness in Drosophila 35,36,37. Importantly, we found that oral administration of a sub-dose of the TH inhibitor alpha-methyl-p-tyrosine (AMPT) fully rescued the total sleep amount in Rogdi mutants (Fig. 4b). AMPT also significantly lengthened ABL in Rogdi mutants but not in control flies, indicating that AMPT treatment substantially restored sleep consolidation in Rogdi mutants. Conversely, L-DOPA administration shortened the total sleep amount and ABL in control flies but not in Rogdi mutants (Fig. 4c), indicating that Rogdi mutants are more resistant to the wake-promoting effects of L-DOPA. At the dosages used in our experiments, both AMPT and L-DOPA affected sleep latency comparably in control flies and Rogdi mutants (Supplementary Fig. 5).
A dopaminergic pathway mediates the wake-promoting effects of Rogdi mutation. (a) A schematic of a dopaminergic synapse. AMPT, alpha-methyl-p-tyrosine (an inhibitor of tyrosine hydroxylase); DA, dopamine; DAT, dopamine transporter; DopR, D1-like dopamine receptor; L-DOPA, L-3,4-dihydroxyphenylalanine (a precursor of DA); TH, tyrosine hydroxylase. (b) Oral administration of AMPT rescues short baseline sleep in Rogdi mutants. Sleep behaviors in individual flies were analyzed similarly to the data presented in Fig. 1. Gray and orange bars indicate wild-type (w 1118) and Rogdi del mutant backgrounds, respectively. Data represent average ± SEM (n = 30–63). Two-way ANOVA detected significant interaction between Rogdi mutation and AMPT effects on sleep amount (F[1,155] = 71.13, P < 0.0001) and average sleep bout length (ABL) (F[1,155] = 9.102, P = 0.0030). (c) Rogdi mutation desensitizes the wake-promoting effects of L-DOPA (n = 34–63). Two-way ANOVA detected significant interaction between Rogdi mutation and L-DOPA effects on sleep amount (F[1,185] = 8.59, P = 0.0038) and ABL (F[1,185] = 10.2, P = 0.0017). (d) Blocking synaptic transmission in dopaminergic (TH > TNT, tetanus toxin light chain) but not octopaminergic (TDC2 > TNT) neurons masks the wake-promoting effects of Rogdi mutation (n = 23–80). Two-way ANOVA detected significant interaction of Rogdi mutation with TH > TNT (F[2,230] = 15.42, P < 0.0001) but not with TDC2 > TNT (F[2,191] = 0.1922, P = 0.8253). (e) Hypomorphic mutation in a dopamine receptor gene, dumb, suppresses the short sleep phenotypes in Rogdi mutants (n = 30–126). Two-way ANOVA detected significant genetic interaction between Rogdi and dumb mutations on sleep amount (F[1,280] = 44.72, P < 0.0001) and ABL (F[1,280] = 28.05, P < 0.0001). n.s., not significant, *P < 0.05, ***P < 0.001 to no-drug or heterozygous controls in the same genetic backgrounds as determined by Tukey post hoc test.
To further validate the implication of dopaminergic transmission in Rogdi-dependent sleep regulation, we examined if genetic disruption in the DA pathway could mask Rogdi effects on baseline sleep. Transgenic expression of tetanus toxin light chain (TNT) in TH-expressing neurons could block neurotransmission specifically at the dopaminergic synapses. This genetic manipulation indeed suppressed the wake-promoting effects of Rogdi mutation (Fig. 4d, Supplementary Fig. 6). By contrast, synaptic blockade of the wake-promoting octopaminergic transmission24 did not affect the short sleep phenotypes in Rogdi mutants, verifying the specific involvement of the dopaminergic circuit in Rogdi-dependent sleep. In addition, hypomorphic mutation in the D1-like DA receptor dumb 35, 36 also rescued Rogdi mutant phenotypes (Fig. 4e, Supplementary Fig. 7). Collectively, our pharmacological and genetic data demonstrate that the wake-promoting DA pathway mediates the sleep suppression observed in Rogdi mutants.
GABAergic Transmission via Metabotropic GABA Receptors Supports Rogdi-dependent Sleep
Gamma-aminobutyric acid (GABA) is known to be involved in sleep regulation and its sleep-promoting effects are well conserved between flies and mammals17. We thus assessed possible roles of the inhibitory neurotransmitter GABA in the wake-promoting effects of Rogdi mutation (Fig. 5a). Mitochondrial GABA transaminase (GABA-T) metabolizes GABA into succinic semialdehyde to suppress GABAergic transmission38. A low dose of the GABA-T inhibitor, ethanolamine O-sulfate (EOS) modestly elevated sleep amount in control flies, whereas it fully rescued sleep latency and substantially increased sleep amount in Rogdi mutants (Fig. 5b). Since GABA-T expressed in glial cells has been implicated in promoting wakefulness38, we next asked if elevated GABA levels in the synaptic clefts or glia could rescue Rogdi mutant sleep. Synaptic GABA levels are lowered by GABA transporter (GAT) that translocates extracellular GABA back to presynaptic neurons and glial cells, thereby restraining GABAergic transmission39. We found that the short sleep phenotypes in Rogdi mutants were also rescued by the GAT inhibitor, DL-2,4-diaminobutyric acid (DABA) (Fig. 5c). These data support that the pharmacological elevation of GABAergic transmission could mask the wake-promoting effects of Rogdi mutation, suggesting that deficits in the sleep-promoting GABAergic pathway might be responsible for the sleep phenotypes in Rogdi mutants.
Rogdi-dependent sleep involves a sleep-promoting pathway of GABAergic transmission via metabotropic GABA receptors. (a) A schematic of GABAergic synaptic transmission. DABA, DL-2,4-diaminobutyric acid (GAT inhibitor); EOS, ethanolamine O-sulfate (GABA-T inhibitor); GABAA-R, ionotropic GABA receptor; GABAB-R, metabotropic GABA receptor; GABA-T, GABA transaminase; GAD1, glutamate decarboxylase 1; GAT, GABA transporter; SKF-97541, 3-Aminopropyl(methyl)phosphinic acid (GABAB-R agonist); THIP, 4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridin-3-ol (GABAA-R agonist); VGAT, vesicular GABA transporter. (b,c) Oral administration of EOS (b) or DABA (c) rescues sleep quantity and sleep latency in Rogdi mutants. Sleep behaviors in individual flies were analyzed similarly to the data presented in Fig. 1. Gray and orange bars indicate wild-type (w 1118) and Rogdi del mutant backgrounds, respectively. Data represent average ± SEM (n = 108–120 for EOS; n = 36–78 for DABA). Two-way ANOVA detected significant interaction of Rogdi mutation with EOS and DABA effects on sleep amount (F[1,449] = 21.43, P < 0.0001 for EOS; F[1,209] = 55.2, P < 0.0001 for DABA), average sleep bout length (ABL) (F[1,449] = 4.284, P = 0.0390 for EOS only) and sleep latency (F[1,449] = 18.61, P < 0.0001 for EOS; F[1,209] = 16.61, P < 0.0001 for DABA). (d) An agonist of GABAB-R (SKF-97541) but not GABAA-R (THIP) fully rescues the short sleep phenotypes in Rogdi mutants (n = 29–92). Two-way ANOVA detected significant interaction of Rogdi mutation with SKF-97541 effects on sleep amount (F[1,228] = 56.39, P < 0.0001), ABL (F[1,228] = 33.81, P < 0.0001) and sleep latency (F[1,228] = 14.38, P = 0.0002) but not with THIP. n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001 to no-drug controls in the same genetic backgrounds as determined by Tukey post hoc test.
To independently validate this hypothesis, we examined the sleep-promoting effects of GABA receptor agonists40, 41. Oral administration of 4,5,6,7-tetrahydroisoxazolo[5,4-c]pyridin-3-ol (THIP), an agonist of ionotropic GABA receptors, promoted sleep quantity comparably in control and Rogdi mutant flies while it shortened sleep latency in Rogdi mutants (Fig. 5d). By contrast, a low dose of the metabotropic GABA receptor agonist SKF-97541 affected ABL very modestly in control flies while it fully restored normal sleep behaviors in Rogdi mutants. These data suggest that sleep-promoting GABAergic transmission via metabotropic GABA receptors might be prominently compromised by Rogdi mutations. Taken together, our pharmacological and genetic evidence demonstrates that Rogdi-dependent sleep regulation involves two opposing effects of the wake-promoting DA and sleep-promoting GABA pathways.
Rogdi in GABAergic Neurons Acts Upstream of a Wake-promoting Dopaminergic Pathway to Promote Sleep
We next asked if Rogdi expression in either dopaminergic or GABAergic neurons is sufficient to sustain sleep behaviors. Transgenic ROGDI expression in TH-expressing dopaminergic neurons failed to rescue the short sleep phenotypes in Rogdi mutants (Fig. 6a, Supplementary Fig. 8a). However, ROGDI expression in GABAergic neurons, driven by the glutamate decarboxylase 1 (GAD1)-Gal4 transgene42, partially but significantly rescued sleep behaviors in Rogdi mutants (Fig. 6b, Supplementary Fig. 8b). To confirm that Rogdi is endogenously expressed in GABAergic neurons, we examined Rogdi-expressing neurons in adult fly brains. Since our anti-ROGDI antibodies were limited to the detection of endogenous ROGDI proteins in immunoblots, we employed an enhancer trap line that expresses a transgenic Gal4 driver from the Rogdi locus (Rogdi-Gal4), likely reflecting the endogenous expression of ROGDI. Rogdi-Gal4 was broadly expressed in both anterior and posterior regions in the whole-mount brain when its spatial expression was indirectly visualized by green fluorescent proteins (Fig. 6c, Supplementary Fig. 9). Co-immunostaining with anti-GABA antibodies further revealed GABA-positive neurons among other Rogdi-expressing neurons, particularly in the anterior brain.
Rogdi in GABAergic neurons promotes sleep. (a,b) Transgenic ROGDI expression in GABAergic (GAD1 > ROGDI) but not dopaminergic (TH > ROGDI) neurons rescues short sleep in Rogdi mutants. Sleep behaviors in individual flies were analyzed similarly to the data presented in Fig. 1. Gray and orange bars indicate wild-type (w 1118) and Rogdi del mutant backgrounds, respectively. Data represent average ± SEM (n = 24–43). In the dopaminergic rescue, two-way ANOVA detected significant effects of Rogdi mutation (F[1,229] = 220.3, P < 0.0001) but not TH > ROGDI expression (F[2,229] = 0.5346, P = 0.5866). In the GABAergic rescue, two-way ANOVA detected significant effects of Rogdi mutation (F[1,188] = 149.3, P < 0.0001) and GAD1 > ROGDI expression (F[2,188] = 62.12, P < 0.0001) as well as their significant interaction (F[2,188] = 9.253, P = 0.0001). n.s., not significant, **P < 0.01 to both heterozygous controls in the same genetic backgrounds as determined by Tukey post hoc test. (c) Confocal imaging of Rogdi-expressing neurons in a whole-mount brain. Fluorescent signals from nuclear green fluorescent proteins expressed by an enhancer-trapping transgene in the Rogdi locus (Rogdi-Gal4, green) and GABA (magenta) were visualized in the anterior part of the adult fly brain. Bottom panels indicate fluorescence-specific magnifications of the area in the dotted white box. Arrows indicate GABAergic neurons that express Rogdi-Gal4. (d,e) An agonist of GABAB-R (SKF-97541) but not GABAA-R (THIP) suppresses short sleep caused by constitutive excitation of TH-expressing dopaminergic neurons (TH > NaChBac). Data represent average ± SEM (n = 25–50). Two-way ANOVA detected significant interaction of genotypes with the oral administration of SKF-97541 (F[2,225] = 22.67, P < 0.0001) but not THIP (F[2,221] = 1.485, P = 0.2288). ***P < 0.001 to both heterozygous controls in the same genetic backgrounds; # P < 0.05 to TH-Gal4/ + control only as determined by Tukey post hoc test.
Given that Rogdi effects on sleep were suppressed by pharmacological and genetic blockade of DA transmission, we reasoned that the wake-promoting DA pathway may act downstream of the Rogdi-expressing GABAergic neurons via metabotropic GABA receptors. To validate this hypothesis, we first examined if RNA interference-mediated depletion of metabotropic GABA receptors in TH-expressing dopaminergic neurons could phenocopy Rogdi mutation. However, none of the RNAi lines caused short sleep phenotypes comparable to those in Rogdi mutants (Supplementary Fig. 10). While lack of RNAi phenotypes does not necessarily verify the absence of the sleep-promoting metabotropic GABA receptors in dopaminergic neurons, we could not rule out the possibility that the inhibitory GABAergic input to DA pathway might be mediated indirectly by interneurons. To further validate our original hypothesis above, we examined if GABA receptor agonists could suppress short sleep phenotypes caused by the genetic excitation of TH-expressing dopaminergic neurons. Transgenic expression of the bacterial sodium channel NaChBac43 in TH-expressing neurons (TH > NaChBac) potently shortened daily amount of sleep (Fig. 6d,e) although phenotypic difference between Rogdi mutation and constitutive excitation of TH neurons was observed in sleep latency (Supplementary Fig. 11). Nonetheless, the short sleep phenotype was partially but significantly rescued by oral administration of SKF-97541 whereas sleep-promoting effects of THIP were comparable between TH > NaChBac flies and their heterozygous controls. These data suggest that GABAergic transmission via metabotropic GABA receptors could titrate the wake-promoting effects of the DA pathway regardless of their dopaminergic expression as consistent with our model for Rogdi-dependent sleep regulation.
Insomnia-like Behaviors in Rogdi Mutants Are Sensitized to Select Anti-epileptic Drugs
GABA has long been implicated in many aspects of neural dysfunction including seizures10,11,12, a well-penetrated neurological phenotype in KTS patients. Moreover, sleep deficits frequently accompany epilepsy13,14,15,16, and their severity is positively correlated in human epileptic patients and animal models44,45,46. In fact, sleep deprivation can cause or even aggravate epileptic seizures46,47,48. Not surprisingly, many anti-epileptic drugs (AEDs), including those promoting GABAergic transmission, also modulate sleep behaviors15. We therefore asked if GABA-relevant AEDs could restore sleep behaviors in Rogdi mutants.
Gabapentin and valproate are two AEDs that enhance GABAergic transmission and have been shown to ameliorate seizure susceptibility in Drosophila 47, 49, 50. We found that oral administration of valproate but not gabapentin fully rescued Rogdi mutant phenotypes in total sleep amount and sleep latency at a dosage showing negligible effects in control flies (Fig. 7). In contrast, the short sleep phenotypes in Rogdi mutants were aggravated by anticonvulsant carbamazepine (CBZ). A previous study demonstrated that CBZ rather suppresses sleep behaviors in Drosophila and its wake-promoting effects are, in part, attributable to increased sleep latency by the desensitization of Resistant to dieldrin, an ionotropic GABA receptor51. We note, however, that these AEDs could modulate neural activities in a GABA-independent manner. For instance, CBZ is known as a blocker of voltage-gated sodium channels52 whereas mechanisms underlying anticonvulsant effects of gabapentin and valproate still remain elusive. Nonetheless, these data indicate that Rogdi mutant sleep is sensitized to select but not all AEDs relevant to GABAergic transmission and possibly correlate with the observation that seizures in Rogdi-associated KTS patients are often resistant to anti-epileptic drugs53.
Insomnia-like Behaviors in Rogdi Mutants Are Sensitized to Select Anti-epileptic Drugs. Oral administration of anti-epileptic drugs showed differential effects on short sleep phenotypes in Rogdi mutants. Sleep behaviors in individual flies were analyzed similarly to the data presented in Fig. 1. Gray and orange bars indicate wild-type (w 1118) and Rogdi del mutant backgrounds, respectively. Data represent average ± SEM (n = 21–97). Two-way ANOVA detected significant interaction of Rogdi mutation with valproate and CBZ on sleep amount (F[1,185] = 17.73, P < 0.0001 for valproate; F[1,114] = 24.88, P < 0.0001 for CBZ) and sleep latency (F[1,185] = 3.892, P = 0.0500 for valproate; F[1,114] = 4.482, P = 0.0364 for CBZ). No significant interactions were observed between Rogdi mutation and gabapentin. n.s., not significant, *P < 0.05, ***P < 0.001 to no-drug controls in the same genetic backgrounds as determined by Tukey post hoc test.
Discussion
Modeling of neurological diseases and disease-relevant genes has greatly advanced our understanding of the fundamental principles that underlie disease pathogenesis as well as brain function. Here, we established the first genetic model of the KTS-associated disease gene Rogdi to demonstrate that Rogdi functions as a novel sleep-promoting factor in GABAergic neurons by promoting GABA transmission. While GABA-dependent sleep regulation via ionotropic GABA receptors have been well documented in Drosophila 41, 51, 54,55,56, our data suggest that GABAergic transmission via metabotropic GABA receptors might be primarily compromised by Rogdi mutation. Furthermore, we identified the wake-promoting DA pathway as a neural locus downstream of Rogdi-dependent GABA signaling given that Rogdi mutant sleep could be rescued by pharmacological or genetic manipulation of dopaminergic transmission. This sleep-regulatory pathway was further supported by our observation that wake-promoting effects of TH-expressing dopaminergic neurons could be selectively titrated by an agonist of metabotropic GABA receptors. Rogdi thus defines a novel pathway coupling these two neurotransmitters to promote baseline sleep in Drosophila as exemplified in other behavioral paradigms across species57,58,59. On the other hand, Rogdi-dependent GABA transmission might have inhibitory effects on a sleep-promoting neural pathway for sleep homeostasis60, 61 to suppress recovery sleep after sleep loss.
What is the molecular basis by which Rogdi supports GABAergic transmission and promotes sleep? A possible role of ROGDI as a transcription factor has been suggested by the nuclear localization of human ROGDI protein, particularly in the nuclear envelope of blood mononuclear cells and dermal fibroblasts2, and by the conservation of a putatively dimerizing leucine zipper (ZIP) motif among ROGDI homologs5. Several lines of evidence, however, argue against this possibility. bZIP transcription factors possess basic residues followed by their ZIP domains whereas ROGDI protein lacks the canonical motif (i.e., basic residues) for DNA-binding activity and nuclear localization. Drosophila ROGDI actually displays its subcellular distribution in both nucleus and cytoplasm of cultured cells or adult fly neurons (Supplementary Fig. 3) although the exclusive nuclear localization might not be a prerequisite for transcriptional activities. We recently reported the crystal structure of human ROGDI protein62 and showed that, unlike other bZIP transcription factors, human ROGDI protein exists as a monomer containing two structurally distinguishable domains (designated as α and β domains, respectively) (Supplementary Fig. 2a). The α domain exhibits an α-helical bundle that consists of H1, H2, H3, and H6 helices. In fact, the ZIP-like motif in the α domain appears to mediate their intramolecular interactions, contributing to the overall structure and stability of a monomeric ROGDI protein. Based on sequence homology between Drosophila and human ROGDI proteins, we predict that Rogdi[del] allele removes the majority of the first helix including the repeated leucine residues in the α domain and the first three strands in the β domain, explaining the instability of ROGDIdel proteins (Fig. 1c, Supplementary Fig. 2b). A smaller but comparable deletion of the ZIP-like motif has been reported in a KTS patient with a splicing mutation in human Rogdi gene5. In addition, a functional study in cervical cancer cell lines demonstrated Rogdi effects on cell cycle progression and radio-sensitivity9. However, further investigations will be required to understand how these cellular phenotypes could be linked to the molecular and neural function of ROGDI protein.
What will be the relevance of our findings to KTS pathogenesis? Genetic heterogeneity has been reported among KTS patients, indicating that Rogdi-independent genetic mutations could contribute to KTS pathogenesis4, 63. A recent study indeed showed that familial mutations in a sodium-citrate transporter gene SLC13A5 are the second genetic cause of KTS64. The pathogenic phenotypes commonly found in Rogdi- and SLC13A5-associated KTS gives rise to the intriguing possibility that these two genes might work together to control the intracellular levels of citrate65. This idea is further supported by the relevance of citrate metabolism to neurological phenotypes in KTS patients. Neurons are energetically dependent on astrocytes because neurons lack pyruvate carboxylase, an enzyme that converts pyruvate to oxaloacetate in the citric acid cycle66, 67. SLC13A5 plays an important role in the transport of glial citrate into neurons to supplement the neuronal citric acid cycle and thereby supply cellular energy68, 69. Furthermore, citrate, an intermediate in the citric acid cycle, acts as a precursor of α-ketoglutarate, which can be metabolized to glutamate and GABA70, implicating SLC13A5 in the biogenesis of GABA. Consistently, anti-epileptic drugs that elevate GABAergic transmission rescued the seizure phenotypes in SLC13A-associated KTS patients65. In addition, we showed that the pharmacological enhancement of GABAergic transmission by oral administration of GABA-T or GAT inhibitors was sufficient to rescue the short sleep phenotypes in Rogdi mutant flies.
Our genetic studies strongly implicate Rogdi function in GABAergic transmission, providing the first clue to understanding the neurological phenotypes observed in KTS patients. Molecular and neural deficits selectively caused by Rogdi mutation might explain why seizures in Rogdi-associated KTS are often resistant to anti-epileptic drugs53, as exemplified by our pharmacological rescue of Rogdi mutant sleep with a specific AED (Fig. 7). Future studies should thus address if Rogdi mutant flies display seizure-like behaviors similarly as in KTS patients and if Rogdi-dependent neural relay of GABAergic transmission controls seizure susceptibility in parallel with baseline sleep. In addition, it will be important to determine whether sleep deficiencies are also observed in KTS patients and whether reduced GABAergic transmission in Rogdi- and, possibly, SLC13A5-associated KTS patients is responsible for their neural dysfunctions, including early-onset seizures. Taken together, our genetic model would constitute an important platform for elucidating the molecular and neural pathogenesis underlying KTS and hint towards a precise development of a therapeutic strategy for KTS in the future.
Materials and Methods
Fly Stocks
All flies were maintained in standard cornmeal–yeast–agar medium under 12 hour light: 12 hour dark cycles at 25 °C. w 1118 (BL5905), TH-Gal4 (BL8848), TDC2-Gal4 (BL9313), GAD1-Gal4 (BL51630), UAS-NaChBac-EGFP (BL9467), UAS-GABA-B-R1 RNAi (BL28353, BL51817), and UAS-GABA-B-R2 RNAi (BL27699) stocks were obtained from the Bloomington Drosophila Stock Center. dumb 2 (f02676) was obtained from the Exelixis Collection at Harvard Medical School. UAS-TNT was described previously71. Rogdi-Gal4 (C0113) was a gift from J. Dubnau (Cold Spring Harbor Laboratory). Rogdi P1 (12866R-1) was obtained from the National Institute of Genetics, Japan. The imprecise excision of the P element insertion in Rogdi P1 mutants was induced by genetic crosses to a transgenic line expressing a transposase. Excision lines were individually established and screened for large deletions in the genomic Rogdi locus to isolate the Rogdi del allele. Rogdi mutant stocks were isogenized by outcrossing to w 1118 backgrounds more than six times prior to the behavioral analyses. A full-length cDNA encoding ROGDI-PA was PCR-amplified from head cDNA library and inserted into a modified pUAS-C5 with a C-terminal 3xFLAG tag72. The UAS-ROGDI-3xFLAG transgene was then injected into w 1118 to establish several independent UAS-ROGDI lines (BestGene Inc).
Behavioral Analyses
Behavioral data were recorded using the Drosophila Activity Monitor system (Trikinetics) under 12-hour light: 12-hour dark cycles at 25 °C. Each male fly was transferred into a 65 × 5 mm glass tube containing 5% sucrose and 2% agar food. Locomotor activity in individual flies was quantified by counting the number of infrared beam crosses per minute. A sleep bout was defined as a behavioral episode during which flies did not show any activity for 5 minutes or longer. Sleep parameters were analyzed with an Excel macro73. Mechanical sleep deprivation was conducted by Sleep Nullifying Apparatus (SNAP)74 while sleep behaviors in individual flies were continuously monitored on the SNAP device before or after the sleep deprivation. Since Rogdi P1 allele has an unrelated RNA interference transgene and induces its overexpression by Gal4 drivers, most behavioral tests including transgenic rescue experiments were performed in Rogdi del mutant backgrounds.
Drug Treatment
AMPT (Sigma), L-DOPA (Sigma), EOS (Tokyo Chemical Industry), DABA (Sigma), and valproic acid (Sigma) were directly dissolved at the indicated concentrations in the food used during behavioral testing, which contained 5% sucrose and 2% agar (‘behavior food’). THIP (Tocris), SKF-97541 (Tocris), CBZ (Acros), and gabapentin (Sigma) were first dissolved at 10 mg/ml (THIP, SKF-97541, CBZ) or 100 mg/ml (gabapentin) and then diluted in the behavior food. For AMPT treatment, flies were fed on AMPT-containing behavior food for 12 hours in the dark phase then switched to standard behavior food at the beginning of the next day; subsequent sleep behaviors were monitored for 24 hours. The wake-promoting effects of L-DOPA were similarly assessed except that 25 μg/ml of ascorbic acid was also included in the behavior food and sleep behaviors were monitored on the same day during 24-hour administration of L-DOPA. For EOS, CBZ, and DABA treatments, flies were pre-fed on drug-containing behavior food for 3 days (EOS, CBZ) or 1.5 days (DABA) and their sleep behaviors were monitored for 24 hours while continuing to be fed on drug-containing food. For THIP, SKF-97541, gabapentin, and valproate treatments, flies were pre-fed on drug-containing behavior food for 1.5 days and their sleep behaviors were monitored for 3 days (Rogdi mutants and their control flies) or for 24 hours (TH > NaCh and their control flies) while continuing to be fed on drug-containing food.
Western Blot Analysis
Two rabbits were immunized with bacterially purified, full-length Drosophila ROGDI protein to generate polyclonal anti-ROGDI antibodies (AbFrontier). Fly heads were collected and homogenized in a lysis buffer (25 mM Tris-Cl pH 7.5, 300 mM NaCl, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol, 0.5% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride). Protein extracts were resolved by 10% SDS-PAGE, transferred to Protran nitrocellulose membranes (GE Healthcare), and immunoblotted with rabbit anti-ROGDI (diluted at 1:2000) or mouse anti-TUBULIN (diluted at 1:2000, Developmental Studies Hybridoma Bank) antibodies. After overnight incubation with the primary antibodies at 4 °C with gentle agitation, protein blots were further incubated with horseradish peroxidase-conjugated secondary antibodies (diluted at 1:5000, Jackson ImmunoResearch) and detected by Clarity Western ECL blotting substrate (Bio-Rad) using ImageQuant LAS 4000 (GE Healthcare).
Whole-brain Imaging
Adult fly brains were dissected in phosphate-buffered saline (PBS), fixed in PBS containing 3.7% formaldehyde, and then blocked with 0.5% normal goat serum (NGS) in PBS containing 0.3% Tritox X-100 (PBS-T). Dissected brains were incubated with rabbit anti-GABA antibody (diluted in PBS-T containing 0.5% NGS and 0.05% sodium azide at 1:1000, Sigma) for 2 days at 4 °C. After washing with PBS-T, brains were further incubated with anti-rabbit Alexa Flour 594 antibody (diluted at 1:600, Jackson ImmunoResearch) for 1 day at 4 °C, washed with PBS-T, and then mounted in a VECTASHIELD mounting medium (Vector Laboratories). Confocal images were acquired with a Laser Scanning Confocal Microscope (FV1000, Olympus) and analyzed in ImageJ software.
Quantification of Protein Stability
Drosophila S2 cells were cultured in Shields and Sang M3 insect medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (ThermoFisher Scientific) at 25 °C. Expression vectors for either wild-type ROGDI or ROGDIdel protein with a C-terminal 3xFLAG tag were transiently transfected using Effectene Transfection Reagent according to the manufacturer’s instructions (QIAGEN). Transfected cells were further incubated with 100 μg/ml of cycloheximide to block protein synthesis for the indicated hours before their harvest at 48 hours after transfection. Total cell extracts were resolved by SDS-PAGE and immunoblotted with anti-FLAG or anti-TUBULIN antibodies. Intensities from immunoblotting signals were quantified using ImageJ software. Relative levels of ROGDI proteins at each time-point were calculated by normalizing to those of TUBULIN proteins. Fitting curves for the time-dependent decay of ROGDI proteins were generated using Excel and used to estimate their half-life in hours.
Data Availability
All data analyzed during this study are included in this published article (and its Supplementary Information file). The raw datasets generated during the current study are available from the corresponding authors on reasonable request.
References
Kohlschutter, A. et al. Familial epilepsy and yellow teeth–a disease of the CNS associated with enamel hypoplasia. Helv Paediatr Acta 29, 283–294 (1974).
Mory, A. et al. A nonsense mutation in the human homolog of Drosophila rogdi causes Kohlschutter-Tonz syndrome. Am J Hum Genet 90, 708–714, https://doi.org/10.1016/j.ajhg.2012.03.005 (2012).
Schossig, A. et al. Mutations in ROGDI Cause Kohlschutter-Tonz Syndrome. Am J Hum Genet 90, 701–707, https://doi.org/10.1016/j.ajhg.2012.02.012 (2012).
Tucci, A. et al. Kohlschutter-Tonz syndrome: mutations in ROGDI and evidence of genetic heterogeneity. Hum Mutat 34, 296–300, https://doi.org/10.1002/humu.22241 (2013).
Huckert, M. et al. A Novel Mutation in the ROGDI Gene in a Patient with Kohlschutter-Tonz Syndrome. Mol Syndromol 5, 293–298, https://doi.org/10.1159/000366252 (2014).
Mory, A. et al. Kohlschutter-Tonz syndrome: clinical and genetic insights gained from 16 cases deriving from a close-knit village in Northern Israel. Pediatr Neurol 50, 421–426, https://doi.org/10.1016/j.pediatrneurol.2014.01.006 (2014).
Dubnau, J. et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol 13, 286–296 (2003).
Busch, S. J. & Sassone-Corsi, P. Dimers, leucine zippers and DNA-binding domains. Trends Genet 6, 36–40 (1990).
Chen, Y. F. et al. Downregulation of a novel human gene, ROGDI, increases radiosensitivity in cervical cancer cells. Cancer Biol Ther, 1–9, doi:https://doi.org/10.1080/15384047.2016.1219818 (2016).
Treiman, D. M. GABAergic mechanisms in epilepsy. Epilepsia 42(Suppl 3), 8–12 (2001).
Sperk, G., Furtinger, S., Schwarzer, C. & Pirker, S. GABA and its receptors in epilepsy. Adv Exp Med Biol 548, 92–103 (2004).
Khazipov, R. GABAergic Synchronization in Epilepsy. Cold Spring Harb Perspect Med 6, a022764, https://doi.org/10.1101/cshperspect.a022764 (2016).
Derry, C. P. & Duncan, S. Sleep and epilepsy. Epilepsy Behav 26, 394–404, https://doi.org/10.1016/j.yebeh.2012.10.033 (2013).
Manni, R. & Terzaghi, M. Comorbidity between epilepsy and sleep disorders. Epilepsy Res 90, 171–177, https://doi.org/10.1016/j.eplepsyres.2010.05.006 (2010).
Jain, S. V. & Glauser, T. A. Effects of epilepsy treatments on sleep architecture and daytime sleepiness: an evidence-based review of objective sleep metrics. Epilepsia 55, 26–37, https://doi.org/10.1111/epi.12478 (2014).
Giorelli, A. S., Passos, P., Carnaval, T. & Gomes Mda, M. Excessive daytime sleepiness and epilepsy: a systematic review. Epilepsy Res Treat 2013, 629469, https://doi.org/10.1155/2013/629469 (2013).
Sehgal, A. & Mignot, E. Genetics of sleep and sleep disorders. Cell 146, 194–207, https://doi.org/10.1016/j.cell.2011.07.004 (2011).
Andretic, R., Franken, P. & Tafti, M. Genetics of sleep. Annu Rev Genet 42, 361–388, https://doi.org/10.1146/annurev.genet.42.110807.091541 (2008).
Cirelli, C. The genetic and molecular regulation of sleep: from fruit flies to humans. Nat Rev Neurosci 10, 549–560, https://doi.org/10.1038/nrn2683 (2009).
Cirelli, C. et al. Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092, https://doi.org/10.1038/nature03486 (2005).
Kume, K., Kume, S., Park, S. K., Hirsh, J. & Jackson, F. R. Dopamine is a regulator of arousal in the fruit fly. J Neurosci 25, 7377–7384, https://doi.org/10.1523/JNEUROSCI.2048-05.2005 (2005).
Koh, K. et al. Identification of SLEEPLESS, a sleep-promoting factor. Science 321, 372–376, https://doi.org/10.1126/science.1155942 (2008).
Stavropoulos, N. & Young, M. W. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 72, 964–976, https://doi.org/10.1016/j.neuron.2011.12.003 (2011).
Crocker, A. & Sehgal, A. Octopamine regulates sleep in drosophila through protein kinase A-dependent mechanisms. J Neurosci 28, 9377–9385, https://doi.org/10.1523/JNEUROSCI.3072-08a.2008 (2008).
Pfeiffenberger, C. & Allada, R. Cul3 and the BTB adaptor insomniac are key regulators of sleep homeostasis and a dopamine arousal pathway in Drosophila. PLoS Genet 8, e1003003, https://doi.org/10.1371/journal.pgen.1003003 (2012).
Garbe, D. S. et al. Changes in Female Drosophila Sleep following Mating Are Mediated by SPSN-SAG Neurons. J Biol Rhythms 31, 551–567, https://doi.org/10.1177/0748730416668048 (2016).
Isaac, R. E., Li, C., Leedale, A. E. & Shirras, A. D. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc Biol Sci 277, 65–70, https://doi.org/10.1098/rspb.2009.1236 (2010).
Borbely, A. A. & Achermann, P. Sleep homeostasis and models of sleep regulation. J Biol Rhythms 14, 557–568 (1999).
Konopka, R. J. & Benzer, S. Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68, 2112–2116 (1971).
Yu, Q. et al. Molecular mapping of point mutations in the period gene that stop or speed up biological clocks in Drosophila melanogaster. Proc Natl Acad Sci USA 84, 784–788 (1987).
Shaw, P. J., Tononi, G., Greenspan, R. J. & Robinson, D. F. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417, 287–291, https://doi.org/10.1038/417287a (2002).
Dubowy, C. et al. Genetic Dissociation of Daily Sleep and Sleep Following Thermogenetic Sleep Deprivation in Drosophila. Sleep 39, 1083–1095, https://doi.org/10.5665/sleep.5760 (2016).
Andretic, R., van Swinderen, B. & Greenspan, R. J. Dopaminergic modulation of arousal in Drosophila. Curr Biol 15, 1165–1175, https://doi.org/10.1016/j.cub.2005.05.025 (2005).
Wisor, J. P. et al. Dopaminergic role in stimulant-induced wakefulness. J Neurosci 21, 1787–1794 (2001).
Liu, Q., Liu, S., Kodama, L., Driscoll, M. R. & Wu, M. N. Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr Biol 22, 2114–2123, https://doi.org/10.1016/j.cub.2012.09.008 (2012).
Ueno, T. et al. Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat Neurosci 15, 1516–1523, https://doi.org/10.1038/nn.3238 (2012).
Pimentel, D. et al. Operation of a homeostatic sleep switch. Nature 536, 333–337, https://doi.org/10.1038/nature19055 (2016).
Chen, W. F. et al. A neuron-glia interaction involving GABA transaminase contributes to sleep loss in sleepless mutants. Mol Psychiatry 20, 240–251, https://doi.org/10.1038/mp.2014.11 (2015).
Leal, S. M. & Neckameyer, W. S. Pharmacological evidence for GABAergic regulation of specific behaviors in Drosophila melanogaster. J Neurobiol 50, 245–261 (2002).
Berry, J. A., Cervantes-Sandoval, I., Chakraborty, M. & Davis, R. L. Sleep Facilitates Memory by Blocking Dopamine Neuron-Mediated Forgetting. Cell 161, 1656–1667, https://doi.org/10.1016/j.cell.2015.05.027 (2015).
Dissel, S. et al. Sleep restores behavioral plasticity to Drosophila mutants. Curr Biol 25, 1270–1281, https://doi.org/10.1016/j.cub.2015.03.027 (2015).
Ng, M. et al. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron 36, 463–474 (2002).
Nitabach, M. N. et al. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci 26, 479–489, https://doi.org/10.1523/JNEUROSCI.3915-05.2006 (2006).
Malow, B. A. et al. Identification and treatment of obstructive sleep apnea in adults and children with epilepsy: a prospective pilot study. Sleep Med 4, 509–515 (2003).
Vaughn, B. V., D’Cruz, O. F., Beach, R. & Messenheimer, J. A. Improvement of epileptic seizure control with treatment of obstructive sleep apnoea. Seizure 5, 73–78 (1996).
Roundtree, H. M. et al. Orexin Receptor Antagonism Improves Sleep and Reduces Seizures in Kcna1-null Mice. Sleep 39, 357–368, https://doi.org/10.5665/sleep.5444 (2016).
Lucey, B. P., Leahy, A., Rosas, R. & Shaw, P. J. A new model to study sleep deprivation-induced seizure. Sleep 38, 777–785, https://doi.org/10.5665/sleep.4674 (2015).
Ellingson, R. J., Wilken, K. & Bennett, D. R. Efficacy of sleep deprivation as an activation procedure in epilepsy patients. J Clin Neurophysiol 1, 83–101 (1984).
Kuebler, D. & Tanouye, M. Anticonvulsant valproate reduces seizure-susceptibility in mutant Drosophila. Brain Res 958, 36–42 (2002).
Reynolds, E. R. et al. Treatment with the antiepileptic drugs phenytoin and gabapentin ameliorates seizure and paralysis of Drosophila bang-sensitive mutants. J Neurobiol 58, 503–513, https://doi.org/10.1002/neu.10297 (2004).
Agosto, J. et al. Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci 11, 354–359, https://doi.org/10.1038/nn2046 (2008).
Ambrosio, A. F., Soares-Da-Silva, P., Carvalho, C. M. & Carvalho, A. P. Mechanisms of action of carbamazepine and its derivatives, oxcarbazepine, BIA 2-093, and BIA 2-024. Neurochem Res 27, 121–130 (2002).
Schossig, A., Wolf, N. I., Kapferer, I., Kohlschutter, A. & Zschocke, J. Epileptic encephalopathy and amelogenesis imperfecta: Kohlschutter-Tonz syndrome. Eur J Med Genet 55, 319–322, https://doi.org/10.1016/j.ejmg.2012.02.008 (2012).
Chung, B. Y., Kilman, V. L., Keath, J. R., Pitman, J. L. & Allada, R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol 19, 386–390, https://doi.org/10.1016/j.cub.2009.01.040 (2009).
Haynes, P. R., Christmann, B. L. & Griffith, L. C. A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. Elife 4, https://doi.org/10.7554/eLife.03868 (2015).
Parisky, K. M. et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60, 672–682, https://doi.org/10.1016/j.neuron.2008.10.042 (2008).
van Zessen, R., Phillips, J. L., Budygin, E. A. & Stuber, G. D. Activation of VTA GABA neurons disrupts reward consumption. Neuron 73, 1184–1194, https://doi.org/10.1016/j.neuron.2012.02.016 (2012).
Crickmore, M. A. & Vosshall, L. B. Opposing dopaminergic and GABAergic neurons control the duration and persistence of copulation in Drosophila. Cell 155, 881–893, https://doi.org/10.1016/j.cell.2013.09.055 (2013).
Bjordal, M., Arquier, N., Kniazeff, J., Pin, J. P. & Leopold, P. Sensing of amino acids in a dopaminergic circuitry promotes rejection of an incomplete diet in Drosophila. Cell 156, 510–521, https://doi.org/10.1016/j.cell.2013.12.024 (2014).
Donlea, J. M., Pimentel, D. & Miesenbock, G. Neuronal machinery of sleep homeostasis in Drosophila. Neuron 81, 860–872, https://doi.org/10.1016/j.neuron.2013.12.013 (2014).
Liu, S., Liu, Q., Tabuchi, M. & Wu, M. N. Sleep Drive Is Encoded by Neural Plastic Changes in a Dedicated Circuit. Cell 165, 1347–1360, https://doi.org/10.1016/j.cell.2016.04.013 (2016).
Lee, H. et al. The crystal structure of human Rogdi provides insight into the causes of Kohlschutter-Tonz Syndrome. Sci Rep 7, 3972, https://doi.org/10.1038/s41598-017-04120-x (2017).
De Souza, C. M. et al. Kohlschutter-Tonz syndrome in siblings without ROGDI mutation. Oral Health Dent Manag 13, 728–730 (2014).
Schossig, A. et al. SLC13A5 is the second gene associated with Kohlschutter-Tonz syndrome. J Med Genet. https://doi.org/10.1136/jmedgenet-2016-103988 (2016).
Klotz, J., Porter, B. E., Colas, C., Schlessinger, A. & Pajor, A. M. Mutations in the Na(+)/citrate cotransporter NaCT (SLC13A5) in pediatric patients with epilepsy and developmental delay. Mol Med 22, https://doi.org/10.2119/molmed.2016.00077 (2016).
Shank, R. P., Bennett, G. S., Freytag, S. O. & Campbell, G. L. Pyruvate carboxylase: an astrocyte-specific enzyme implicated in the replenishment of amino acid neurotransmitter pools. Brain Res 329, 364–367 (1985).
Yu, A. C., Drejer, J., Hertz, L. & Schousboe, A. Pyruvate carboxylase activity in primary cultures of astrocytes and neurons. J Neurochem 41, 1484–1487 (1983).
Bergeron, M. J., Clemencon, B., Hediger, M. A. & Markovich, D. SLC13 family of Na(+)-coupled di- and tri-carboxylate/sulfate transporters. Mol Aspects Med 34, 299–312, https://doi.org/10.1016/j.mam.2012.12.001 (2013).
Thevenon, J. et al. Mutations in SLC13A5 cause autosomal-recessive epileptic encephalopathy with seizure onset in the first days of life. Am J Hum Genet 95, 113–120, https://doi.org/10.1016/j.ajhg.2014.06.006 (2014).
Bak, L. K., Schousboe, A. & Waagepetersen, H. S. The glutamate/GABA-glutamine cycle: aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem 98, 641–653, https://doi.org/10.1111/j.1471-4159.2006.03913.x (2006).
Sweeney, S. T., Broadie, K., Keane, J., Niemann, H. & O’Kane, C. J. Targeted expression of tetanus toxin light chain in Drosophila specifically eliminates synaptic transmission and causes behavioral defects. Neuron 14, 341–351 (1995).
Lim, C. & Allada, R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 340, 875–879, https://doi.org/10.1126/science.1234785 (2013).
Pfeiffenberger, C., Lear, B. C., Keegan, K. P. & Allada, R. Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc 2010, prot5520, https://doi.org/10.1101/pdb.prot5520 (2010). pdb.
Seugnet, L. et al. Identifying sleep regulatory genes using a Drosophila model of insomnia. J Neurosci 29, 7148–7157, https://doi.org/10.1523/JNEUROSCI.5629-08.2009 (2009).
Acknowledgements
We thank J. Dubnau, Bloomington Drosophila Stock Center, and the National Institute of Genetics for Drosophila strains. This work was supported by a grant from the Korea Health Technology R&D Project through the KHIDI funded by the Ministry of Health & Welfare, the Republic of Korea (HI16C1747) (C.Lim); a grant from the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MSIP), the Republic of Korea (NRF-2016R1E1A2914795) (C.Lim); a grant from the NRF funded by the MSIP, the Republic of Korea (NRF-2016R1A2B4011111) (J.C.).
Author information
Authors and Affiliations
Contributions
C.Lim and J.C. conceived the study and supervised the work. M.K., D.J., E.Y., Y.O., J.Y.S., J.L., Y.K. and C.Lim generated transgenic flies, performed genetics experiments, and analyzed behavioral data. C.Lee purified ROGDI protein for the generation of anti-ROGDI antibodies. M.K. and E.Y. validated anti-ROGDI antibodies and performed immunoblottings. M.K., D.J., Y.O., J.L., and C.Lim performed immunofluorescence assays. D.J., H.J.S., and O.H. performed biochemical analyses in brain extracts. M.K. and C.Lim wrote the manuscript with input from E.Y. J.Y.S. and J.C. made editorial changes to the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kim, M., Jang, D., Yoo, E. et al. Rogdi Defines GABAergic Control of a Wake-promoting Dopaminergic Pathway to Sustain Sleep in Drosophila . Sci Rep 7, 11368 (2017). https://doi.org/10.1038/s41598-017-11941-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-11941-3
This article is cited by
-
The Rogdi knockout mouse is a model for Kohlschütter–Tönz syndrome
Scientific Reports (2024)
-
Genome-wide association in Drosophila identifies a role for Piezo and Proc-R in sleep latency
Scientific Reports (2024)
-
Perampanel effectiveness in treating ROGDI-related Kohlschütter-Tönz syndrome: first reported case in China and literature review
BMC Medical Genomics (2023)
-
Cleavage of human tau at Asp421 inhibits hyperphosphorylated tau induced pathology in a Drosophila model
Scientific Reports (2020)
-
Sleep, brain vascular health and ageing
GeroScience (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.