Abstract
The molecular analysis of sex in vertebrates is important, as it has the potential to provide vital information for theoretical and applied research alike. Teleost fish are the ancient vertebrates that present a broad sex chromosome system but lack differentiated sex chromosomes in most species. Hence understanding the sex in fish would not only illuminate the sex determination evolution in vertebrates but also shed light on fish farming. In the present study, we used grass carp as a teleost fish model, studied the Y chromosome by using a pool-and-sequence strategy in combination with fragment-ratio method. In total, we identified five Y-linked scaffolds (totaling 347 Kb) and six Y-specific sequences that could be used as sex-specific markers, demonstrating the suitability of NGS-based re-sequencing of pooled DNAs for the identification of sex markers in fish. Moreover, 14 putative Y-linked genes were described for the first time. All the genes, except for un-y1, un-y2, and ubq-y, showed high similarity to their female homologs. RT-PCR revealed that ubq-y was only expressed in the male hypothalamus and pituitary. These findings provided an abundant resource for the Y chromosome of grass carp, and may help elucidate sex chromosome evolution in cyprinid fish.
Similar content being viewed by others
Introduction
Sex determination and differentiation are important issues in biology research. Teleost fish are the ancient vertebrates that present a broad sex chromosome system but lack differentiated sex chromosomes in most species. Hence, studying the evolution of sex chromosomes in teleost is challenging1. Understanding the sex determination mechanisms in fish would provide insights into the sex determination evolution in vertebrates2. In many species, economic traits, such as growth rate, weight, and disease resistance, have been associated with sex3. Therefore, understanding the sex determination mechanisms in farmed fish is necessary. It is urgent to develop sex manipulation biotechnologies for sex control breeding4, 5.
In recent vertebrates and most fish species, sex is mainly determined by one or more genetic factors located in sex chromosomes or autosomes6. The two common sex chromosome systems in most species are XX/XY and ZZ/ZW. However, some species present other types, such as XX/XO type in Giant freshwater stingray (Himantura dalyensis)7, X1X1X2X2/X1X2Y type in Neotropical gymnotiformes electric fish (Gymnotiformes)8, ZZ/ZW1W2 type in Apareiodon hasemani eigenmann (Characiformes: Parodontidae)9 and so on10. Compared with recent vertebrates, fish lack morphologically and genetically differentiated sex chromosomes. Moreover, there are frequent recombinations between X chromosome and Y chromosome. Until now, the studies that focused on sex-specific regions and genes in fish remain rare, thus the sex-specific markers and sex-related genes must be identified to elucidate the complicated mechanisms underlying various sex determination types.
Traditionally, fish geneticists adopt multiple molecular marker technologies to screen sex-specific DNA markers, such as amplified fragment length polymorphic DNA, single nucleotide polymorphisms, random amplified polymorphic DNA, simple sequence repeats and quantitative trait locus11,12,13,14. These DNA markers could help locate sex-related genes. DM-related Y-specific gene (dmy), the representative sex-determining gene, was identified in Medaka (Oryzias latipes) through the traditional strategy15, 16. However, traditional experiments is expensive and time consuming, its screening scale is fairly limited. Therefore, the efficiency of identifying sex-specific markers could still be improved. The advent of high-throughput data and the development of bioinformatics methods have made identification of sex-specific region possible at a genomic scale. Y-linked regions and genes are being screened in more and more species recently17,18,19,20. Hall et al. identified six novel Y-linked genes in Mosquitos (Aedes aegypti) through Illumina sequencing of the male and female genomes basing on previously assembled genomes, one distant homolog of transformer-2 gene, named nix, was screened and proven to be the male-determining gene in Mosquitos17, 21. Meanwhile, Bidon et al. compared two in silico approaches and found that the method based on differences in the average read depth of autosomal versus sex chromosomal scaffolds was more efficient than that was based on similarity searching of known Y-linked genes19. With the former method, a 1.9 Mb region of Y chromosome was identified from the Polar bear (Ursus groenlandicus) genome. These studies demonstrated the reliability of the sequencing read depth approach that sequenced the pools of male and female individuals separately. As a result, the region wherein only male reads are aligned is regarded as the candidate sequence for the Y chromosome. However, the above approaches must be based on previous male genome assemblies.
Grass carp (Ctenopharyngodon idellus) is an important freshwater aquaculture cyprinid fish worldwide, and its genome has been recently sequenced through whole-genome shotgun sequencing in a gynogenetic female adult and a wild water-captured male adult grass carp respectively22. Comparative analysis of the zebrafish genome revealed that the LG24 linkage group of grass carp corresponds to chromosomes 10 and 22 of zebrafish, suggesting that the grass carp genome underwent chromosome fusion during evolution. Therefore, grass carp is an interesting model to study the evolution of sex chromosomes in fish. Our previous work adopted comparison of assemblies between one male and one female, and found one out of 206 contigs that demonstrated male specific amplification finally. The efficiency is still relatively low that may result from individuals’ difference. Hence in the present study, basing on our previous male grass carp genome data, we tested the suitability and reliability of the sequencing read depth approach of pooled DNAs in the identification of sex markers in fish. Finally, we computationally identified 347 Kb Y-linked sequences and 14 Y-linked genes. Six Y-linked sequences are unique in males, demonstrating a higher efficiency. Moreover, one Y-linked gene ubiquitin ligase gene (ubq-y) presents a male-specific expression, further studies of this gene may help elucidate its roles in sex determination and differentiation. Our findings will provide valuable insights into the evolution of fish sex chromosome.
Results
Sexual proportion statistics
A total of 90 grass carp individuals (18 months old) were selected randomly from the full-sib population and dissected to observe the gonad development under the microscope. Micrographs of the gonads are shown in Fig. 1. The result of gender identification revealed a sex ratio close to 1:1 with 47 females and 43 males. Moreover, the 90 grass carp individuals from the gynogenetic population were all females. The sexual proportion statistics confirmed that grass carp has a heteromorphic XX/XY sex chromosome system. Females, featuring two X chromosomes, are the homogametic sex, whereas males, featuring one X and Y chromosome each, are the heterogametic sex.
Preliminary data analysis of re-sequencing of pooled DNAs
A total of 30 female and 30 male grass carp individuals at 18 months old were selected from the full-sib population. DNA was extracted, and two DNA mixed pools were constructed, hereafter referred to as female-pool and male-pool. The male-gc-assembly were softmasked using RepeatMasker and split using the repeat region as a separator. Consequently, 605,448 fragments were retained with a size of 749 Mb and an N50 value of 1,760 bp. The sizes of clean data from the female-pool and the male-pool were 33.33 Gb and 32.93 Gb, including 222,208,685 and 219,531,391 reads, respectively. The statistics of reference-guided mapping is shown in Table 1. Although the number of total reads from the female-pool was higher than that from the male-pool, the mapped reads (153,321,131) of the former was less than that of the latter (159,341,024), and mapping rates corresponded to 69% and 72.58%, respectively. The higher mapping rate of the male-pool suggested the existence of male-specific sequences in the reference that cannot be sequenced in the female-pool. The overall process for identifying Y-linked sequences and genes in the grass carp genome is shown in Supplementary Fig. S1.
Identification of putative Y-linked scaffolds
The fragment-ratio method was applied to identify Y-linked fragments. We introduced several abbreviations in this method as shown in Materials and Methods. Fi-reads is the number of female-pool aligned reads against Fragmenti in male-gc-assembly, and Mi-reads is the number of male-pool aligned reads against Fragmenti in male-gc-assembly. Thus, Ri denotes the ratio that divides the Fi-reads by the Mi-reads, Ri-norm is the normalization of Ri value, and can fairly describe the difference in the ratio of alignment reads against the reference genome between the female-pool and the male-pool. The average Ri-norm for all the fragments was 0.905 (Supplementary Fig. S2). We set a threshold of Ri-norm <0.3 to differentiate Y-linked fragments from autosome and X fragments. Considering that false positives could result from low coverage, we excluded fragments with Mi-reads less than 15. Finally, 923 fragments from 0 to 0.3 were considered as putative Y-linked fragments, involving 107 scaffolds with a size of 1,121 Kb. Of the 923 fragments, 169 have Ri-norm values below 0.15. Information for these fragments is shown in the Supplementary Table S1.
Since scaffolds from Y chromosome is composed of a number of Y-linked fragments, then it is reasonable that scaffolds from Y chromosome tend to have higher rate of Y-linked fragments when compared with scaffolds from autosome. Hence, an enrichment analysis was performed on the 107 scaffolds following the formula that were described in Materials and Methods. Only the P-values below 1e-005 were supposed to be significantly enriched. Finally, six out of 107 scaffolds were detected to have significantly more fragments with Ri-norm values less than 0.3 (P-value < 1e-005). These six scaffolds are Sca3194, Sca704, Sca713, Sca28791, Sca811, and Sca971 (Table 2). Take Sca971 for example, the length of Sca971 is about 45,330 bp, a total of 28 fragments were produced, interestingly, 18 out of 28 fragments have the Ri-norm values below 0.3, while 10 fragments have the Ri-norm values below 0.15, the P-value of Sca971 is 2.166655e-44. Conversely, for putative autosome-linked scaffolds, such as Sca667, rare putative Y-linked fragments (2/78) were detected, and its P-value was 0.006456602 that above 1e-005. The discrepancies in Ri-norm distribution for scaffolds that originated from different chromosomes are shown in Fig. 2. Compared with autosome-linked scaffolds (Sca0), putative Y-linked scaffolds have a lower Ri-norm value (~0). Conversely, putative X-linked scaffolds (Sca628) have a higher Ri-norm value (~2). Thus, the six putative scaffolds, with a combined size of 356 Kb, were more likely to be from the Y chromosome.
Discrepancy of Ri-norm distribution among fragments that originated from different chromosomes. Pink circle indicates Ri-norm values of fragments. The x axis represents different fragments, and the y axis represents Ri-norm value. Scaffolds are shown by their ID numbers. Compared with autosome-linked scaffolds that with Ri-norm value of ~1, Y-linked scaffolds have lower Ri-norm value (~0), whereas X-linked scaffolds have higher Ri-norm value (~2).
High abundance of repetitive elements on the Y-linked scaffolds
For the six putative Y-linked scaffolds, we analyzed their repeat contents. Compared with whole male genome, 17.53% of the bases were masked as repetitive elements, the repetitive elements rate of Y-linked scaffold reached up to 30.71%. The detailed information for the repetitive elements and gaps is listed in Supplementary Tables S3 and S4. Moreover, we found a remarkably higher abundance of retro elements on the Y-linked scaffolds. The coverage of the retro elements is nearly five times as much sequences compared with the entire genome (17.16% vs. 3.53%). Additionally, the average coverage of long interspersed nuclear elements on the Y-linked scaffolds was 10.43%, which is 10 times more than that of the whole genome (1.34%). Furthermore, more long-terminal-repeat elements, L2/CR1/Rex, interspersed and simple repeats were detected in the Y-linked scaffolds. Notwithstanding, similar rates of DNA transposons (10.41% vs. 12.03%) were present in both Y-linked scaffolds and entire genome.
PCR tests in of full-sib population confirmed Y-linked sequences
We selected a batch of putative Y-linked sequences from the six scaffolds for further PCR verification in the full-sib population. To improve the efficiency of this process, we chose fragments that have higher stringent threshold (Ri-norm < 0.15). Thus, 75 fragments were included for subsequent analysis. A BlASTN search23 was performed against female-gc-assembly using default parameters22, and fragments with best blast hits (expect value < 1e-005) were discarded. Finally, 17 sequences across five scaffolds (except Sca3194) were retained in PCR amplification in the full-sib population. The detailed information is listed in Supplementary Table S2. PCR amplification was performed to test the accuracy of our approach. Fragments were defined as Y-linked when a single, clear amplification product was detected in males, whereas no amplicons or only low-intensity bands of different sizes were observed in females. Full-sib PCR results showed that most fragments with Ri-norm less than 0.15 were amplified only in males with an accuracy of 94.12% (16/17) (Supplementary Table S2). Partial full-sib PCR results of representative sequences for the five scaffolds are shown in Table 3 and Fig. 3. Therefore, the five scaffolds were totally confirmed as Y-linked. The above results suggested that fragments with low Ri-norm values tend to be Y chromosome sequences, and the fragment-ratio method is reliable for identifying Y-linked sequences in grass carp.
PCR tests for five representative Y-linked fragments in full-sib grass carp individuals. All five fragments that distributed in five significantly enriched scaffolds (Sca971, Sca811, Sca713, Sca28791, and Sca704) were selected in PCR tests in eight female and male grass carp samples. These five sequences are Sca971_3_662, Sca713_52_382, Sca28791_1_303, Sca811_22_1407, Sca704_77_319. Male specificity was defined as the occurrence of a clear amplicon of a distinct size in males but not in females. The DL2000 DNA Markers are loaded in the Lanes. The result verified that these five putative Y-linked scaffolds were Y-linked. Additional information of these five representative Y-linked fragments can be found in Table 3.
PCR tests in wild population confirmed Y-specific sequences
Differences in the genetic background among different families may cause sequence divergence for the Y chromosome and affect test efficiency. Therefore, eight female and eight male wild grass carp individuals from different water systems (Zhujiang river, Yangtse river, Xiangjiang river, Lao river.) that were described in our previous study24, were collected. The detection of polymorphism in those wild grass carps confirmed that the natural germplasm resources of wild grass carp were genetically diverse24. Further PCR tests were performed in these wild grass carps to confirm that whether the Y-linked sequence is specific to wild male individuals. However, the results of further PCR in wild grass carps were below our expectations (Supplementary Fig. S3). All six sequences of Sca971 (Sca971_3_662, Sca971_30_188, Sca971_4_1894, Sca971_9_1908, and Sca971_32_446, Sca971_34_3238) still presented male-specific amplification (see Fig. 4). These sequences have the potential to be used as sex-specific molecular markers. By contrast, the 11 other sequences (Sca 704_15_1909, Sca 704_33_2332, Sca 704_63_267, Sca 704_77_319, Sca 713_22_716, Sca 713_52_382, Sca 713_68_619, Sca 713_93_3485, Sca 811_22_1407, Sca 811_50_718, Sca 28791_1_303) from other scaffolds could be amplified in certain wild females, as shown in Supplementary Table S2.
PCR tests for fragments that distributed in Sca971 in wild grass carp individuals. Male specificity was defined as the occurrence of a clear amplicon of a distinct size in males but not in females. The DL2000 DNA Markers are loaded in the Lanes. The results showed that all the six sequences of Sca971 were only amplified in wild male grass carps.
Y-linked genes annotation
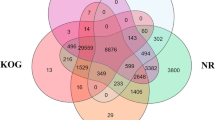
Identification of Y-linked sequences is important to elucidate the gene content of Y chromosomes. The five confirmed Y-linked scaffolds (Sca704, Sca713, Sca28791, Sca811, and Sca971) were used for gene annotation based on BLAST evidence. Alignments detected in at least one database (NR, Ref-Seq, EST) were considered Y-linked genes. Thus, four scaffolds presented hits in at least one database. A total of 14 genes were annotated from BLAST evidence and were named according to their putative functions or as ‘un-y’ for the genes without previously described functions. Detailed information of all Y-linked genes is listed in Table 4 and Fig. 5. We identified three copies of neoverrucotoxin subunit beta gene (nsb-y1, nsb-y2, and nsb-y3) that located in Sca704 and Sca811. These genes contain Fibronectin type-III, neoverrucotoxin subunit domains and were predicted to play roles in microtubule organization and stabilization25. Furthermore, two copies of the NACHT, LRR and PYD domains-containing gene (nacht-y1 and nacht-y2) were identified in Sca704 and Sca713, they were supposed to function in innate immunity and inflammation26. RNA-directed DNA polymerase genes (rdp-y1, rdp-y2, rdp-y3, and rdp-y4) were widespread in all of these four scaffolds and may play roles in DNA replication and respond to genotoxic stress27. Two copies of retrotransposon-like gene (retro-y1 and retro-y2) were identified in Sca70428. One gene that supposed to be an E3 ubiquitin ligase containing a Zinc finger domain29, named ubq-y gene, was identified in Sca811. All the above genes had Ref-Seq evidence and shared homology with proteins with known functions. In addition, nucleotide BLAST results against the grass carp annotated genes showed that all the genes have female homologs with high sequence identities except ubq-y gene. The coding sequence of ubq-y is obviously divergent from its female homolog. Two other genes were detected in Sca713 and Sca971, nucleotide BLAST results showed that they only have EST evidences and share no similarity to proteins with known functions. We named them unknown function genes (un-y1, un-y2), these two genes could be novel genes because of their lack of similarity to known function genes. The BLAST results showed that many genes are incomplete and some of them lack exons, it is understandable because Y-linked sequences tend to have sequence assembly errors resulting from highly repetitive sequences scattered in the Y chromosome. Therefore, re-sequencing and rapid amplification of complementary DNA are usually needed to fully annotate Y-linked genes.
Predicted gene models of Y-linked scaffolds (Sca971, Sca811, Sca713, and Sca704). Numbers above gene structures indicate predicted start/end position of protein translation in the scaffold. Directions of translation are indicated by black arrowheads. Black boxes with different length represent predicted exons of gene.
Expression analysis of four Y-linked genes
Most Y-linked genes showed high similarity to female homologs, testing male-specific expression is difficult owing to the high homologous sequence. Hence, we only chose four genes, including three genes (ubq-y, un-y1, un-y2) that lack highly similar female homologs, and one gene rdp-y4 that have highly similar female homolog. Expression experiments were performed in four female and male tissues (B: brain, H: hypothalamus, G: gonad, P: pituitary). The result is shown in Fig. 6. We found that rdp-y4 gene was amplified in all four tissues in both sexes with high abundance, thereby confirming the quality of the cDNA template. Hence, rdp-y4 can be used as the positive control. Noticeably, the ubq-y mRNA was highly expressed in two male tissues (hypothalamus and pituitary), but not detected in the female tissues. Expression specificity in male neuroendocrine tissues indicated that ubq-y may function in sex differentiation. By contrast, the un-y1 and un-y2 genes were not amplified in both sexes, suggesting that they may be expressed during developmental stages, such as embryonic or nymphal stage.
Expression of two Y-linked genes (rdp-y4, ubq-y) in females and males. Template cDNAs were derived from four tissues (B: brain, H: hypothalamus, G: gonad, and P: pituitary). Each tissue was extracted from three adult male/female individuals. rdp-y4 gene was amplified in all four tissues in both sexes with high abundance, thereby confirming the quality of the cDNA templates. ubq-y was not detected in the female tissues and was highly expressed in two male tissues (hypothalamus and pituitary).
Considering that the ubq-y gene is only present in male grass carp, we compared its translated coding sequence with female homolog. Twenty four amino acids in N-terminal were failed to be predicted. Alignment results showed widespread discrepancies in amino acid sites (Fig. 7a), suggesting that ubq-y is a divergent paralog (amino acid identity of 75%). Phylogenetic analyses of ubq-y genes suggested that this gene diverged from its female homologous gene in certain fish species, such as Sinocyclocheilus rhinocerous (Fig. 7b), which is also a ray-finned fish in the Cyprinidae family. By contrast, no ubq-y ortholog was detected in zebrafish despite being in the same Cyprinidae family. These findings indicated that their divergence from zebrafish followed by independent divergence of the sex chromosomes in these two species. Both expression and sequence evidences revealed the divergent function of ubq-y gene.
Phylogenetic analysis of ubq genes and alignment of grass carp ubq-y gene with its female homolog. (a) Alignment of coding protein sequences between ubq-y gene and its female homolog in grass carp. Sequences were aligned with ClustalW2. The result showed widespread differences in amino acid sites, suggesting that ubq-y gene is a divergent paralog (amino acid identity of 75%). (b) Phylogenetic analysis of ubq genes. A Maximum Likelihood tree with Poisson correction was constructed using MEGA646. Bootstrap support values (10,000 replicates) are shown inside the tree. The tree is drawn to scale, with branch lengths measured using the number of substitutions per site (below the branches). All positions with gaps and missing data have been eliminated. Sequence accession numbers are shown in Supplementary Table S7. Phylogenetic analyses of ubq genes suggested that ubq-y gene diverged from the female homologous gene in certain fish species. Species information are: Sinocyclocheilus rhinocerous (Srh); Astyanax mexicanus (Ame); Cyprinodon variegatus (Cva); Oryzias latipes (Ola); Dictyostelium discoideum (Ddi); Homo sapiens (Hum); Danio rerio (Dre).
Discussion
The Y chromosome is involved in multiple biological functions, such as male fertility and sex determination. A male-determining gene in the Y chromosome is hypothesized to initiate male sexual differentiation, such as Sex-determining region Y gene (sry) in Human and dmy in Medaka15, 30. However, deciphering gene content and organization is difficult because of the highly heterochromatic nature and accumulation of repetitive DNA in sex chromosomes, especially the Y chromosome. Fish are known to have moderately differentiated sex chromosomes31, 32. Recombination events between the X and Y chromosomes are frequent, thereby complicating the characterization of sex-specific fish sequences and genes. Nevertheless, considering the diverse sex-determination types and evolutionary position, studying sex chromosomes in fish is valuable and important to evolutionary biology. Grass carp is an interesting model for sex chromosomes because of its sex chromosome fusion event described in our previous work22.
With the advent of high-throughput sequencing and reduced cost, various types of sequencing strategies were applied in acquisition of objective traits, including whole genome re-sequencing, restriction site associated DNA sequencing (RAD-seq), exome sequencing33, 34 and so on. Despite of higher cost, whole genome re-sequencing (WGS) demonstrates its advantage of higher coverage compared with other next-generation sequencing (NGS) technologies. Whole genome re-sequencing of pools (Pool-seq) is becoming an efficient and economical way to acquire the objective traits, and applied in identifying markers linked to any specific gene or genomic region35. In our study, we performed whole genome re-sequencing of pooled gDNA samples of both sexes in grass carp. A fragment-ratio method was successfully applied to identify five Y-linked scaffolds (totaling 347 Kb), with a 94.12% accuracy of PCR tests in full-sibs (16/17). This method is based on the discrepancy in the ratio of alignment reads against the male reference genome between the female-pool and the male-pool. Thus, it could work in large and complex genomes and can be applied to any species with differentiated sex chromosomes. Coincidentally, numerous Y-linked genes have been successfully described following the same principle in various species, such as the Kissing bug (Triatoma dimidiata), Polar bear, and Mosquitos17,18,19. Our study further confirmed the efficacy of such comparative genomic strategies in fish. Compared with traditional methods, such as AFLP and SNP, these comparative genomic strategies are more efficient and time saving, especially in detecting large Y-linked region and genes.
However, the species previously studied have highly evolved gonosomes, and the prominent divergence between sex chromosomes make it very easy to acquire Y-specific sequences and genes. In contrast, identifying Y-specific genes in fish is complicated by their primitive property accompanied by the high sequence similarity of gonosomes. Although the method we used follows the same principle of the CQ method proposed by Hall et al.32, 36, it has a higher rate of false positivity in PCR tests in the wild grass carp population compared with Mosquitoes. Only 6 out of 17 Y-linked sequences were proven male specific in the wild grass carp population. The undifferentiated status of fish gonosomes and high frequent crossovers between X and Y chromosome likely explain much of this phenomenon. For Y chromosome, the high similarity to X chromosome counterpart make it difficult to distinguish from each other through a pair of primer. Our results indicated the crucial influence affected by discrepancy of gonosomes. The higher the similarity between X and Y chromosome, the lower the chance of finding true Y-specific sequences and genes. Notwithstanding, we still screened out six Y-specific sequences (spanning 48Kb) that could be used as sex-specific markers with a higher efficiency compared with our previous work22, where only one out of 206 contigs retained its specificity in wild grass carps finally. Actually, the putative male-specific markers (probe 184, CI01M090293) that was described in our earlier publication22, was located on Sca971 around the 30 Kb position (30,957–34,697 bp), with a length of 4,148 bp. Additionally, the amplified product of the specific marker corresponds to Sca971_34_3238, which belongs to our six Y-specific sequences, was also verified by our PCR test in both full-sib and wild grass carp individuals (Supplementary Table S2.xls).
Placental mammals have highly evolved sex chromosomes, but lower recombination with other genome regions make the Y chromosome genetically degenerate with rare active genes36. Fish still have a primitive Y chromosome that evolved from recent evolutionary events, thus, its gene content is theoretically higher than that of placental mammals. Although we proved that the Y-linked scaffolds have a higher rate of repetitive elements compared with the whole genome (30.71% vs. 17.53%), 14 genes were still deciphered on the basis of homology searching. Except for un-y1, un-y2, and ubq-y genes, the Y-linked genes showed high similarities to their female homologs, most of them are multi-copy genes distributed in at least two loci. This phenomenon revealed that degeneration level of Y chromosome in fish is still primitive despite of accumulation of repetitive DNA. For Sca971, although we have confirmed the male specificity of six sequences, all of them spread across an intergenic region, the gene content of this scaffold still shares high sequence similarity to female homologs, such as rdp-y4 gene. These findings indicated that the extent of divergence is variable across the region, with high similarity in coding sequences but low similarity in intergenic regions that result from mutation or indels with few functional constraints37.
We have no indication of the biological function of the two other Y-linked genes (un-y1 and un-y2) because of the lack of similarity to genes with known functions. ubq-y gene was supposed to be an E3 ubiquitin-protein ligase containing a zinc finger domain and shared low sequence similarity to its female homolog. RT-PCR results further revealed that ubq-y is expressed only in the male hypothalamus and pituitary, suggesting ubq-y gene possibly participates in gender differentiation and development in grass carp. Future functional studies, such as RNAi silencing, are needed to clarify its biological role. The alignments and phylogenetic analysis showed the extensive sequence divergence of ubq-y with female homologs, suggesting the differentiation of sex chromosomes in certain cyprinid fish. Although the grass carp genome underwent whole-genome duplication similar to zebrafish after the teleost radiation, with its adaption to variable environments, grass carp undergone species-specific duplications involving cell proliferation and differentiation, as well as organ size control22, which make it higher in terms of sex differentiation. Grass carp already has incomplete differentiation of the Y chromosome compared with zebrafish, gene duplication and function divergences are accompanied by the evolution of sex chromosomes.
Materials and Methods
Ethical procedures
Animal welfare and experimental procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006), and the study protocol was approved by the Committee of the Institute of Hydrobiology, Chinese Academy of Sciences (CAS). All surgeries were performed under eugenol anesthesia, all efforts were made to minimize suffering.
Construction of population and experimental fish collection
During the breeding season of grass carp and common carp (late April to early May), 1 female grass carp, 1 male grass carp, and 1 male common carp parents were randomly selected from the Guanqiao Experimental Station of the Institute of Hydrobiology, CAS (Wuhan, China). Semen or eggs from each fish were collected separately. A part of eggs were conventionally fertilized using grass carp semen to construct a full-sib population. Another part of eggs were fertilized using UV-inactivated semen of common carp and simulated in cold water at 0–4 °C to construct a gynogenetic population38. The two groups of fertilized eggs were hatched in different hatchery ponds for emerging fry. After 2 weeks, 500–800 fry with approximately 1 month old were selected randomly from each population and bred in two ponds approximately 2600 m2 in area.
At age of 18 months, weighing 800–1,200 g, with the average length of 30 cm, 90 grass carp individuals were selected randomly from each population and were dissected to observe gonad development under the microscope. In addition, eight female and eight male wild grass carp individuals were collected from different wild water systems (Zhujiang river, Yangtse river, Xiangjiang river, Lao river) that were described in our previous study. The GPS coordinates of the five water systems are listed in Supplementary Table S8. The detection of polymorphism in those wild grass carps confirmed that the natural germplasm resources of wild grass carp were genetically diverse24. Tail fins were cut from experimental fish and preserved in 95% ethanol.
The fish were fed in the Guan Qiao Experimental Station, Institute of Hydrobiology, Chinese Academy of Sciences, and acclimatized in aerated fresh water at 26–28 °C for one week before processing. Fish were fed with a commercial diet twice a day and water was exchanged daily.
Re-sequencing of DNA pools of male and female grass carp
A total of 30 female and 30 male grass carp individuals at 18 months old were selected from the full-sib population. DNA was extracted from tail fins using the conventional phenol–chloroform method. Thus, two DNA mixed pools were constructed, hereafter referred to as female-pool and male-pool. DNA quality and purity were checked using 1% agarose gel electrophoresis and ultra-micro spectrophotometry (Nanodrop 2000, USA). A total of 1.5 μg DNA per sample was used as input material for the DNA library. Sequencing libraries were generated using TruSeqNano DNA HT Sample Preparation Kits (Illumina, USA) following the manufacturer’s protocol (NEBNext Ultra DNA Library Prep Kit for Illumina), index codes were added to attribute sequences to each sample. The DNA sample was disrupted via sonication into fragments of approximately 500 bp in size. The DNA fragments were end-polished, A-tailed, and ligated using the full-length adapter for Illumina sequencing with further PCR amplification. Finally, PCR products were purified, libraries were analyzed for size distribution using an Agilent2100 Bioanalyzer and quantified using real-time PCR. These libraries were sequenced using the Illumina HiSeq4000 platform, 150 bp paired-end reads were generated with an insert size of approximately 500 bp. Library constructions and sequencing were performed using Novogene Company (China). The sequencing data in this study have been deposited in the Sequence Read Archive (SRA) at the National Center for Biotechnology Information (NCBI) (accession number: SRP095438).
Preliminary data analysis
The quality control of paired-end Illumina sequencing data was initially evaluated using the NGSQC Toolkit, and low-quality sequence data were filtered out (cutOffQualScore < 20)39. The grass carp genome assemblies were obtained from our previous work22. The female genome was referred to as female-gc-assembly, the male genome assembly that was utilized as the reference genome source was referred to as male-gc-assembly. Male-gc-assembly is arranged into 130,221 scaffolds with an N50 value of 2,269 Kb and a size of 1.07 Gb, its average sequence length is approximately 8,265 Kb, with the minimum and maximum at 246 bp and 16.34 Mb, respectively. The scaffolds in male-gc-assembly were labeled according to their respective scaffold ID numbers with the format “ScaID.” In consideration that repetitive sequences may result in a high bias of sequencing coverage, all scaffolds from male-gc-assembly were softmasked using RepeatMasker and split using the repeat region as a separator. Fragments with length shorter than 200 bp were disregarded.
RepeatMasker40 was used to softmask all scaffolds and avoid repetitive sequences (i.e., transposable elements, virus retrogenes, and simple repeats) using default parameters, except that the “-species” parameter was set as “Dario”. As a result, scaffolds in male-gc-assembly were fragmented into numerous fragments. Meanwhile, fragments shorter than 200 bp were excluded to avoid false positives. The remaining sequences were used as reference.
The treated cleaned reads were aligned to against the reference genome using the ultrafast read aligner bwa41. Samtools42 was used to index, merge, sort, remove, format convert, and remove duplications against the aligned data. Picard (http://picard.sourceforge.net/) was used to mark duplicated reads, and realignment of reads around regions of indels was performed using GATK43.
Computational identification of putative Y-linked sequences
The fragment-ratio method was applied to identify Y-linked sequences, this method was based on the discrepancy in the ratio of alignment reads against the reference genome (male-gc-assembly) between the female-pool and the male-pool. For a given Fragmenti, we can obtain Ri-norm that was calculated using the following equations:
where Fi-reads is the number of female-pool aligned reads against Fragmenti in male-gc-assembly; Mi-reads is the number of male-pool aligned reads against Fragmenti in male-gc-assembly; Ri denotes the ratio that divides the Fi-reads by the Mi-reads. However, the discrepancy in the sequencing coverage of the two pools may affect the Ri value, leading to a biased result. Hence, Ri normalization was performed by multiplying by the value of norm, which denotes the normalized female-male ratio that was calculated by dividing Ftotal-reads from Mtotal-reads. Therefore, Ri-norm can fairly describe the difference in the ratio of alignment reads against the reference genome between female-pool and male-pool. The depth statistics of all the fragments was calculated using the command “idxstats” in Samtools. Moreover, a program written in perl named “RatioOfSex.pl” was used to calculate the Ri-norm value.
The fragment-ratio method is based on the difference in the relative numbers of X and Y chromosomes between females and males. Reads from the female-pool and the male-pool demonstrate the characteristics of sex-specific patterns when aligned against different chromosomes. For species with the XX/XY sex determination system, the female has two X chromosomes and no Y chromosome, whereas the male has one X and Y chromosome each. Moreover, both sexes carry two copies of each autosome. Thus, unique Y-linked sequences are not present in the female genome. On the basis of the above data, the values of Ri-norm for Fragmenti vary from 0 to 2: ~0 for Y chromosome fragments, ~1 for autosome fragments, and ~2 for X chromosome fragments. Meanwhile, some Y-linked fragments have homologous regions to their X chromosome counterparts, leading to few aligned reads from the female-pool, hence Ri-norm is greater than 0. We set a threshold of Ri-norm as 0.3 to distinguish Y-linked sequences from autosomes and X chromosomes as previously described17. False positives could arise from low coverage, thus the resulting fragments with Mi-reads less than 15 were excluded from further analysis.
Hypergeometric test in R was used to perform enrichment analysis for the Y-linked scaffolds following the below equation:
where N and n are the total number of fragments and the total number of fragments with Ri-norm below the threshold (<0.3) in the reference genome, respectively. M and k represent the total number of fragments and the number of fragments below the threshold (<0.3) in the scaffold, respectively. Scaffolds with P-value < 1e-005 were used as putative Y-linked scaffolds.
Selection of putative Y-linked fragments and PCR verification
We selected a batch of fragments for further PCR verification. To improve this process, for the fragments with Ri-norm below the threshold (<0.15), BLASTN search23 was performed against female-gc-assembly using default parameters22, fragments with the best BLAST hits (expect value < 1e-005) were discarded. The remaining putative Y-linked fragments were used for PCR amplification in eight male and eight female grass carp individuals (18 months old) randomly selected from a full-sib population. To determine whether the Y-linked sequences are specific to the wild population, we also performed PCR tests in eight male and eight female wild grass carp individuals (more than 18 months old) from different water systems. DNA was extracted from tail fins using the conventional phenol-chloroform method. The primers are listed in Supplementary Table S7. The PCR reaction system was 20 μL in volume, the PCR program was as follows: initial denaturation at 94 °C for 2 min, denaturation at 94 °C for 30 s, annealing at 51 °C for 30 s, and extension at 72 °C for 30 s. After 39 cycles, the program was extended at 72 °C for 2 min. Primers were designed using Primer5 and synthesized by Sangon Company (Shanghai, China). Y-linked sequences were defined as the occurrence of a clear amplicon or a distinct size in males but not in females.
Y-linked genes annotation
The Y-linked scaffolds that were successfully validated by PCR tests in the full-sib population were used for Y-linked gene annotation. These scaffolds were softmasked and blasted against different databases to search for gene coding regions. Non-redundant proteins (NR), reference sequence proteins (Ref-Seq), and expressed sequence tags (EST) were downloaded from NCBI separately. Grass carp annotated genes were downloaded from the official National Center for Gene Research website (http://www.ncgr.ac.cn/grasscarp/). BLASTX was used against NR and Ref-Seq (e-value = 1e-005), BLASTN was used against the EST database and grass carp annotated genes (e-value = 1e-005). Alignments detected in at least one database were considered as Y-linked genes, which were then annotated from BLAST evidence and named according to their putative function or ‘un-y’ for the genes without previously described functions. Finally, all Y-linked genes were blasted against the Protein Family of Domains database44.
Expression analysis of four Y-linked genes
Expression experiments were performed in both male and male tissues for the Y-linked genes that lack high similarity to their female homologs. Three male and three female grass carp individuals (18 months old) were selected from the full-sib population. Brain, hypothalamus, gonad, and pituitary samples were prepared, equal quantities of the three samples from the same tissue were mixed, resulting in eight samples. RNA was isolated with Oligotex mRNA Kit (QIAGEN, Germany) in accordance with the manufacturer’s protocol. Primers were designed using Primer5 and synthesized by Sangon Company (Shanghai, China). The primers are listed in Supplementary Table S7. The PCR reaction system was 20 μL in volume, and the PCR program was as follows: initial denaturation at 94 °C for 2 min, denaturation at 94 °C for 30 s, annealing at 51 °C for 30 s, and extension at 72 °C for 30 s. After 39 cycles, the program was extended at 72 °C for 2 min. Protein sequences were aligned using ClustalW245, and a Maximum Likelihood tree with Poisson correction was constructed using MEGA646.
References
Kobayashi, Y., Nagahama, Y. & Nakamura, M. Diversity and Plasticity of Sex Determination and Differentiation in Fishes. Sex Dev 7, 115–125 (2013).
Devlin, R. H. & Nagahama, Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture 208, 191–364 (2002).
Morita, S. H., Morita, K. & Nishimura, A. Sex-biased dispersal and growth in sablefish (Anoplopoma fimbria) in the northeastern Pacific Ocean. Environ Biol Fish 94, 505–511 (2012).
Weber, G. M. & Lee, C. S. Current and future assisted reproductive technologies for fish species. Adv Exp Med Biol 752, 33–76 (2014).
Martinez, P. et al. Genetic architecture of sex determination in fish: applications to sex ratio control in aquaculture. Front Genet 5, 340 (2014).
Kikuchi, K. & Hamaguchi, S. Novel sex-determining genes in fish and sex chromosome evolution. Dev Dyn 242, 339–353 (2013).
de Souza Valentim, F. C., Porto, J. I., Bertollo, L. A., Gross, M. C. & Feldberg, E. XX/XO, a rare sex chromosome system in Potamotrygon freshwater stingray from the Amazon Basin, Brazil. Genetica 141, 381–387 (2013).
Cardoso, A. L., Pieczarka, J. C. & Nagamachi, C. Y. X1X1X2X2/X1X2Y sex chromosome systems in the Neotropical Gymnotiformes electric fish of the genus Brachyhypopomus. Genet Mol Biol 38, 213–219 (2015).
Bellafronte, E., Schemberger, M. O., Artoni, R. F., Filho, O. M. & Vicari, M. R. Sex chromosome system ZZ/ZW in Apareiodon hasemani Eigenmann, 1916 (Characiformes, Parodontidae) and a derived chromosomal region. Genet Mol Biol 35, 770–776 (2012).
Heule, C., Salzburger, W. & Bohne, A. Genetics of sexual development: an evolutionary playground for fish. Genetics 196, 579–591 (2014).
Gamble, T. et al. Restriction Site-Associated DNA Sequencing (RAD-seq) Reveals an Extraordinary Number of Transitions among Gecko Sex-Determining Systems. Mol Biol Evol 32, 1296–1309 (2015).
Kovacs, B., Egedi, S., Bartfai, R. & Orban, L. Male-specific DNA markers from African catfish (Clarias gariepinus). Genetica 110, 267–276 (2000).
Rao, H. O., Deng, J. C., Wang, W. M. & Gao, Z. X. An AFLP-based approach for the identification of sex-linked markers in blunt snout bream, Megalobrama amblycephala (Cyprinidae). Genet Mol Res 11, 1027–1031 (2012).
Felip, A., Martinez-Rodriguez, G., Piferrer, F., Carrillo, M. & Zanuy, S. AFLP Analysis Confirms Exclusive Maternal Genomic Contribution of Meiogynogenetic Sea Bass (Dicentrarchus labrax L.). Mar Biotechnol 2, 301–306 (2000).
Matsuda, M. et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417, 559–563 (2002).
Matsuda, M. et al. The sex determining gene of medaka: a Y-specific DM domain gene (DMY) is required for male development. Fish Physiol Biochem 28, 135–139 (2003).
Hall, A. B. et al. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. Bmc Genomics 14, 273 (2013).
Koerich, L. B. et al. First report of Y-linked genes in the kissing bug Rhodnius prolixus. Bmc Genomics 17, 100 (2016).
Bidon, T., Schreck, N., Hailer, F., Nilsson, M. A. & Janke, A. Genome-Wide Search Identifies 1.9 Mb from the Polar Bear Y Chromosome for Evolutionary Analyses. Genome Biol Evol 7, 2010–2022 (2015).
Carvalho, A. B. & Clark, A. G. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res 23, 1894–1907 (2013).
Hall, A. B. et al. SEX DETERMINATION. A male-determining factor in the mosquito Aedes aegypti. Science 348, 1268–1270 (2015).
Wang, Y. et al. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nature genetics 47, 625–631 (2015).
Gambin, A. & Wojtalewicz, P. CTX-BLAST: context sensitive version of protein BLAST. Bioinformatics 23, 1686–1688 (2007).
Huang, R. et al. Genetic variations of body weight and GCRV resistance in a random mating population of grass carp. Oncotarget 6, 35433–35442 (2015).
Ueda, A. et al. Purification, properties and cDNA cloning of neoverrucotoxin (neoVTX), a hemolytic lethal factor from the stonefish Synanceia verrucosa venom. Biochim Biophys Acta 1760, 1713–1722 (2006).
Haneklaus, M., O’Neill, L. A. & Coll, R. C. Modulatory mechanisms controlling the NLRP3inflammasome in inflammation: recent developments. Curr Opin Immunol 25, 40–45 (2013).
Sugiyama, T., Cam, H., Verdel, A., Moazed, D. & Grewal, S. I. S. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. P Natl Acad Sci USA 102, 152–157 (2005).
Sekita, Y. et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nature genetics 40, 243–248 (2008).
Niwa, J. et al. A novel centrosomal ring-finger protein, dorfin, mediates ubiquitin ligase activity. Biochem Bioph Res Co 281, 706–713 (2001).
Goodfellow, P. N. & Lovell-Badge, R. SRY and sex determination in mammals. Annu Rev Genet 27, 71–92 (1993).
Charlesworth, D., Charlesworth, B. & Marais, G. Steps in the evolution of heteromorphic sex chromosomes. Heredity (Edinb) 95, 118–128 (2005).
Mank, J. E. & Avise, J. C. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev 3, 60–67 (2009).
Gamble, T. Using RAD-seq to recognize sex-specific markers and sex chromosome systems. Mol Ecol 25, 2114–2116 (2016).
Ramirez-Gonzalez, R. H. et al. RNA-Seq bulked segregant analysis enables the identification of high-resolution genetic markers for breeding in hexaploid wheat. Plant Biotechnol J 13, 613–624 (2015).
Michelmore, R. W., Paran, I. & Kesseli, R. V. Identification of Markers Linked to Disease-Resistance Genes by Bulked Segregant Analysis - a Rapid Method to Detect Markers in Specific Genomic Regions by Using Segregating Populations. P Natl Acad Sci USA 88, 9828–9832 (1991).
Charlesworth, B. & Charlesworth, D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci 355, 1563–1572 (2000).
Charlesworth, B. Sex determination: primitive Y chromosomes in fish. Curr Biol 14, 745–747 (2004).
Zhang, H. et al. Induced gynogenesis in grass carp (Ctenopharyngodon idellus) using irradiated sperm of allotetraploid hybrids. Mar Biotechnol 13, 1017–1026 (2011).
Patel, R. K. & Jain, M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. Plos One 7, 30619 (2012).
Saha, S., Bridges, S., Magbanua, Z. V. & Peterson, D. G. Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res 36, 2284–2294 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43, 11 10 11–33 (2013).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44, 279–285 (2016).
Hung, J. H. & Weng, Z. Sequence Alignment and Homology Search with BLAST and ClustalW. Cold Spring Harb Protoc 2016, 88 (2016).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution 30, 2725–2729 (2013).
Yang, J. et al. The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC biology 14, 1 (2016).
Davidson, W. S. et al. Sequencing the genome of the Atlantic salmon (Salmo salar). Genome Biol 11, 403 (2010).
Meinhardt, L. W. et al. Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases. Bmc Genomics 15, 164 (2014).
Xu, B. et al. Generation and analysis of ESTs from the grass carp. Ctenopharyngodon idellus. Anim Biotechnol 21, 217–225 (2010).
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 31130055), the Strategic Pilot Science and Technology Projects (A) Category, and the Chinese Academy of Science (No. XDA08030203).
Author information
Authors and Affiliations
Contributions
A.Z., R.H., and Y.W. conceived and designed the experiments. A.Z. performed data analysis. R.H., L.C., and L.X. performed the PCR experiments. L.H., Y.L., L.L., and Z.Z. contributed to fish sampling. A.Z., R.H., and Y.W. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, A., Huang, R., Chen, L. et al. Computational identification of Y-linked markers and genes in the grass carp genome by using a pool-and-sequence method. Sci Rep 7, 8213 (2017). https://doi.org/10.1038/s41598-017-08476-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-08476-y
This article is cited by
-
Sex determination mechanisms and sex control approaches in aquaculture animals
Science China Life Sciences (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.