Abstract
To investigate the metabolic biomarkers of predicting the transition from pre-diabetes (pre-DM) to normal glucose regulation (NGR) and diabetes (DM) in a longitudinal cohort study. 108 participants with pre-DM were followed up for ten years and divided into 3 groups according to different glycemic outcomes. 20 participants progressed to DM, 20 regressed to NGR, and 68 remained at pre-DM. Alterations in plasma metabolites in these groups were evaluated by untargeted ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS). Twenty three metabolites related to glycerophospholipid metabolism, oxidation and antioxidation were associated with the process from pre-DM to NGR, while twenty two metabolites related to amino acid metabolism, glycerophospholipid metabolism and mitochondrial β-oxidation played important roles in the progression to DM. Results from stepwise logistic regression analysis showed that five biomarkers (20-Hydroxy-leukotriene E4, Lysopc(20:4), 5-methoxytryptamine, Endomorphin-1, Lysopc(20:3)) were good prediction for the restoration to NGR, and five biomarkers (Iso-valeraldehyde, linoleic acid, Lysopc(18:1), 2-Pyrroloylglycine, Dityrosine) for the development of DM. The findings suggest that the combination of these potential metabolites may be used for the prognosis of pre-DM. Targeting the pathways that involved in these prognostic biomarkers would be beneficial for the regression to NGR and the early prevention of DM among pre-DM.
Similar content being viewed by others
Introduction
Pre-diabetes (pre-DM), typically defined as blood glucose levels above normal but below diabetes thresholds, has been increasing globally and has a high chance of developing diabetes mellitus (DM)1. It is estimated that there were 318 million adults suffering from impaired glucose tolerance in the world by 20152. As to the Chinese adult population, a cross-sectional survey in 2010 reported that the estimated prevalence of pre-DM was 50.1%3. Previous studies4 have shown that about 5–10% of pre-DM progressed to DM every year, and persistent hyperglycemia leads to the complications that are the major source of morbidity, mortality, and cost. Nowadays, There is a common conception that this natural history is not inevitable5. Randomized controlled trials showed that lifestyle intervention or glucose-lowering medications could delay and even reverse the natural course of pre-DM6,7,8. The American Diabetes Association (ADA) recommends at least annual screening via testing fasting plasma glucose (FPG), 2-hr 75 g oral glucose tolerance test (OGTT), or hemoglobin A1c (HbA1c) in those with pre-DM9. However, clinicians are far from satisfied with the presently used method of monitoring glycemic level because of its hysteresis. FPG, OGTT and HbA1c are used more as a diagnostic than a predictive marker. Interestingly, multiple cross-sectional and prospective cohort studies have revealed the metabolism of impaired branched-chain amino acid (BCAA), aromatic amino acid (AAA), free fatty acid (FFA), acylcarnitines and glycerophospholipid are associated with insulin resistance, and many metabolites were considered as biomarkers for the prediction of pre-DM and DM10,11,12,13. However, little is known about the potential metabolic biomarkers of different glycemic prognoses among subjects with pre-DM. More importantly, building a metabolic model for predicting the transition from pre-DM to NGR or DM would be helpful for the early prevention and treatment among individuals with pre-DM.
Metabolomics provides a snapshot of the metabolic dynamics that reflects the response of living systems to pathophysiological stimuli and/or genetic modifications and surrounding environment. Furthermore, in many ways, tanscriptomic, genomic, and proteomic changes are upstream of the final physiology of cells, whereas the metabolic profile is likely closer in response to the disease process14. Ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS) of biofluids can easily detect hundreds of individual species in a single clinical sample, reflecting the biochemical fingerprint of the organism15. Characterized by sensitivity and high mass accuracy, the technique has been employed in identifying novel biomarkers for cancers16,17, metabolic disorders18,19,20, drug toxicity and function21,22, and so on.
With the current study, we characterized the metabolic profiles of fasting plasma samples among the 108 pre-DM at baseline with different outcomes ten years later utilizing untargeted UPLC-QTOF-MS analysis. 23 and 22 metabolites were identified as biomarkers for transition to NGR and DM from pre-DM, respectively. And the underlying biochemical pathways leading to different prognoses were investigated.
Results
Demographic and Clinical Characteristics
108 participants with pre-DM from a longitudinal cohort study were followed up for ten years and were divided into 3 groups according to different glycemic outcomes. 20 participants progressed to DM, 20 regressed to NGR, and 68 remained at pre-DM, respectively.
At baseline, there were no significant differences in ages, gender, body mass index, blood glucose, lipid profile, blood pressure as well as general health conditions among these 3 groups (Supplemental Table S1). At the end-point of the study, no significant differences in the biochemical characteristics were found among the three groups, except for fasting glucose, 2-h glucose and HbA1c (Table 1).
Quality control
The robustness and stability of the method was assessed by repeat analysis of a representative pooled quality control (QC) sample during sample runs. The overlapped total ion current chromatograms of the QC sample demonstrated the repeatability of our UPLC-QTOF-MS system (Supplemental Fig. S1A). The principal component analysis (PCA) performed on QC and other groups revealed that QC samples were clustered in the PCA scores plot (Supplemental Fig. S2A). The percentage coefficient of variation (CV%) of peak intensity was estimated as 4.1–18.6%. These results collectively indicated good repeatability, reliability, and stability of this method for metabolite analysis.
Plasma metabolite profile and makers for NGR and DM
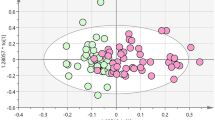
Representative base peak intensity (BPI) chromatograms of plasma samples indicated that the sample metabolites attained suitable separation. Typical single UPLC-QTOF/MS base peak intensity chromatograms of a healthy control, a patient with pre-DM and a patient with DM are presented in Supplemental Fig. S1B. Multivariate statistical analysis was performed to determine whether the plasma metabolic profiles were different among participants progressed to DM, regressed to NGR and remained at pre-DM. The PCA score plots of the three groups, NGR vs DM, NGR vs pre-DM and pre-DM vs DM are presented in Supplemental Fig. S2B–D. The R2X values of PCA analysis were > 0.5 (0.651, 0.604, 0.631, respectively). Partial least squares discriminant analysis (PLS-DA) results of pair-wise groups indicated separations in the three groups (Fig. 1A) and sub-comparison groups (Fig. 1B–D) with valid model fits (R2Y(cum) > 0.7 and Q2(cum) > 0.4)23. Variable importance in projection (VIP) values were obtained from the models and pair-wise statistical test for difference was performed. Variables with VIP value > 1.0 and statistical p value < 0.05 were selected and verified by loading plots. Finally, a metabolite was annotated according to MS information, structure information, accurate mass, retention time, fragmentation pattern and standards.
PLS-DA score plots of different groups based on plasma spectral data of UPLC-QTOF-MS positive ion mode. One point stands for one subject. (A) PLS-DA score plot of the NGR vs pre-DM vs DM groups. (B) PLS-DA score plot of the NGR vs DM groups. (C) PLS-DA score plot of the NGR vs Pre-DM groups. (D) PLS-DA score plot of the pre-DM vs DM groups.
Based on the above steps, a panel of 23 (Table 2) metabolites distinctively discriminated NGR and DM groups as well as NGR and pre-DM groups with a same trend, which were considered to be the potential metabolic biomarkers for the prognosis to NGR. Similarly, a panel of 22 (Table 3) metabolites discriminated NGR and DM groups as well as DM and pre-DM groups with a same trend were considered to be metabolic markers for the prognosis to DM. Compared to the pre-DM group, 18 biomarkers were up-regulated and 5 biomarkers were down-regulated in the NGR group, while 5 biomarkers increased and 17 biomarkers declined in the DM group. Of the substances that were screened in this study, several substances appear to be unknown. We will attempt to identify these unknown substances in future studies.
Metabolic pathways
Pathway analysis carried out by IPA software revealed that three metabolic pathways including glycerophospholipid metabolism, Lipoate Biosynthesis and Incorporation II, and Melatonin Degradation II were found to contribute in the process from pre-DM to NGR (Fig. 2A). While L-carnitine Biosynthesis, Superpathway of Methionine Degradation, Mitochondrial L-carnitine Shuttle Pathway, and Choline Degradation I were associated with the development of DM (Fig. 2B).
Biological network and canonical pathways related to the identified metabolites in NGR (A) and DM (B). Molecules are represented as nodes, and the biological relationship between two nodes is represented as a line. Red symbols represent up-regulated metabolites; green symbols represent down-regulated metabolites. The solid lines and dotted lines show direct and indirect functional relationships, respectively.
Verification and Optimization of Potential Biomakers
Receiver operating characteristic (ROC) curves for each potential biomarker were established to test the probability of ‘single biomarkers’. Area under curve (AUCs) and their CV% values in QC samples are showed in Supplemental Tables S2 and S3. To assess how multiple metabolites collectively classify the NGR/DM and the Pre-DM groups, we built logistic regression model using stepwise selection on the 23/22 metabolites for the samples, respectively.
(A) ROC analysis for discrimination of pre-DM and NGR groups by logistic regression model combining 20-Hydroxy-leukotriene E4, Lysopc(20:4), 5-methoxytryptamine, Endomorphin-1, Lysopc(20:3). (B) ROC analysis for discrimination of pre-DM and DM groups by logistic regression model combining Iso-valeraldehyde, linoleic acid, Lysopc(18:1), 2-Pyrroloylglycine, Dityrosine.
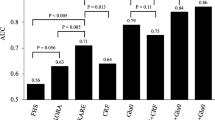
From pre-DM to NGR, five metabolites entered the multiple regression model: 20-Hydroxy-leukotriene E4 (standardized [std] β = 2.135, p = 0.004), Lysopc(20:4) (std β = −1.423, p = 0.027), 5-methoxytryptamine (std β = 0.606, p = 0.047), Endomorphin-1 (std β = 4.685, p = 0.018),and Lysopc(20:3) (std β = −19.176, p = 0.002) with an overall correct percentage of 86.0%. We calculated the sensitivity and specificity based on estimates of the final model built on the samples (logitP1 = −1.847 + 2.135(20-Hydroxy-leukotriene E4) − 1.423(Lysopc(20:4) + 0.606 (5-methoxytryptamine) + 4.685 (Endomorphin-1) − 19.176 (Lysopc(20:3)), and the model fit very well (AUC = 0.910, 95% CI [0.838, 0.983], sensitivity = 84.2%, specificity = 91.0%) with cut-off value −0.08 and p value < 0.001.
Meanwhile, five metabolites entered the multiple regression model from pre-DM to DM: Iso-valeraldehyde (standardized [std] β = −1.544, p = 0.049), linoleic acid (std β = 2.194, p = 0.019), Lysopc(18:1) (std β = −4.769, p = 0.013), 2-Pyrroloylglycine (std β = −2.922, p = 0.044) and Dityrosine (std β = 0.285, p = 0.007) with an overall correct percentage of 95.4%. The final model (logitP2 = −1.119–5.144(Iso-valeraldehyde) + 2.194(linoleic acid) − 4.769(Lysopc(18:1) − 2.922(2-Pyrroloylglycine) + 0.285 (Dityrosine)) showed satisfactory fitness (AUC = 0.976, 95% CI [0.943, 1.000], sensitivity = 90.0%, specificity = 98.5%) with cut-off value −0.33 and p value < 0.001. The ROC curves of the combined biomarkers are shown in Fig. 3. Both of them were statistically different from single metabolites as they showed a higher lower bound of 95% CI of AUCs than most of the upper bounds of 95% CI of single metabolites, and were further validated by comparison of AUCs with MedCalc Statistical Software with a statistical p value < 0.0524.
Relative concentrations of these metabolites in the logistic regression equations are presented in Fig. 4. These data strongly support the robustness of UPLC-QTOF-MS to identify metabolic differences in the plasma samples of pre-DM patients with different prognoses (NGR or DM).
Discussion
Early diagnosis in hyperglycemia, prior to developing into diabetes, can improve the living quality of those suffering from pre-DM. The rapid rise in pre-DM worldwide raises urgent needs to develop effective prognostic biomarkers and prognosis evaluation methods based on the easily accessible materials, such as blood and urine.
Using UPLC-QTOF-MS based metabolic profiling combined with pattern recognition techniques on plasma samples, we identified molecular markers that discriminate the prognoses to NGR and to DM in 108 pre-diabetic patients in a longitudinal study with more than ten years of follow-up. In this study, a total of 23 metabolites involved in different biochemical metabolic pathways with high statistical significance were associated with the outcome of NGR, while 22 metabolites with the outcome of DM.
Of these perturbed metabolic pathways, particular importance should be given to glycerophospholipid metabolism. Earlier studies have demonstrated that diabetes is intimately associated with metabolic disorders of lipids, especially phospholipids25,26. Phosphocholine metabolites, including PC and LysoPC, are key components of the biomembranes of cells, as well as participating in various biological pathways27,28, especially in cellular signaling and metabolism. Correspondingly, significant changes in phosphatidylcholine metabolism were observed in our study. PC (16:0/14:0) and PC (18:0/18:2) showed a reduction in level from pre-DM to DM, while PC (14:1/16:1) increased and PC (18:3/20:3) decreased from pre-DM to NGR. This is in line with a previous prospective cohort study based on 866 participants with seven years’ follow-up that a series of PC such as C32:1, C36:1, C38:3, and C40:5 were independently associated with increased risk of T2D and C34:3, C40:6, C42:5, C44:4, and C44:5 with decreased risk29. However, it was also demonstrated that the type of linkage between phospholipid core and fatty acid residue may be the key factor contributing to the antithetical association between two phosphatidylcholine subclasses and T2DM risk29. Therefore, the specific structural formula of the PCs detected needs further validation.
What’s more, some studies have reported that changed concentrations of lysoPCs are associated with the risk of T2DM, especially lysoPC(18:2), which was significantly altered in patients with impaired glucose tolerance (IGT) and was identified as an IGT-specific biomarker30. LysoPCs can mediate many cell-signaling pathways in monocytes/macrophages29,31 and specific receptors32, and therefore participate in the inflammatory response. In our results, several lysoPC species (LPC C20:4, LPC C18:3, LPC C20:5, LPC C20:3) were down-regulated from pre-DM to NGR group, while the plasma level of LPC C18:1 were higher and LPC C18:0 were lower in DM than pre-DM patients, possibly owing to altered activity of phospholipase A2 which catalyzes PC hydrolysis to lysoPC33. In mouse models, Yea, K et al. have reported that the blood glucose lowering effect of LPC were found to be sensitive to variations in LPC acyl chain length34, which may elucidate the divisive findings mentioned above.
In recent years, 5-methoxytryptamine (5-MT) has been revealed to play a pivotal role in the alternative melatonin synthetic pathway, in which serotonin is first O-methylated to 5-MT and, thereafter, 5-MT is N-acetylated to melatonin (N-acetyl-5-methoxytryptamine)35. Melatonin was reported as a free radical scavenger and an antioxidant for protection from the oxidative stress36,37,38, especially reducing oxidative damage to lipids, which is important for the maintenance of mitochondrial homeostasis. Letra-Vilela, R. et al. observed that removal of the N-acetyl group enhances the antioxidant and neuroprotective properties of the maletonin39. The elevated level of plasma 5-MT concentrations contributed to scavenge radicals and radical products and functioned as antioxidant against reaction oxygen species (ROS). Meanwhile, an increased level of plasma S-(hydroxymethyl)glutathione concentration was observed in patients regressed to NGR. S-(hydroxymethyl)glutathione interconverts with glutathione (GSH), later of which is a key antioxidant and marker for conditions with oxidative stress40. Therefore, we can conclude that a balance between oxidation and antioxidation was achieved in patients regressed to NGR.
Caprylic acid, also named octanoic acid, is a medium-chain fatty acid which down-regulates a number of key adipogenic genes including peroxisome proliferator activated receptor (PPAR), CCAAT/enhancer binding protein alpha. Rats fed on diet rich in medium-chain fatty acids had smaller fat pads, reduced adipose tissue lipoprotein lipase activity and improved insulin sensitivity and glucose tolerance41. Thus the elevated level of caprylic acid may be another contributor to glucose tolerance recovery.
In this study, a major alteration observed in patients progressed to DM was amino acid metabolism with a decreased level of L-lysine, L-threonine, Betaine, Iso-valeraldehyde, 2-ketobutyric acid, and 2-Pyrroloylglycine. 2-Ketobutyric acid is a substance that is involved in the metabolism of many amino acids, such as glycine, methionine, valine, leucine, serine, threonine and isoleucine. It is also one of the degradation products of threonine, which can be converted to propionyl-CoA (and subsequently methylmalonyl CoA, which can be converted to succinyl CoA, a citric acid cycle intermediate), and thus enter the citric acid cycle. Our research is consistent with the report that 2-ketobutyric acid is associated with both prevalent diabetes (OR: 1.43, 95% CI (1.06,1.92)) and incident diabetes (OR: 1.81, 95% CI (1.35, 2.42)42 while glycine is found inversely associated with diabetes risk43,44. This suggest that increased activity in the L-threonine-2-ketobutyric acid-citric acid cycle and decreased glycine, which plays an important role in metabolic regulation and anti-oxidative reactions45 may account for diabetes risk.
Betaine, also glycine betaine, has been reported to be disturbed in diabetes and it is regarded as a marker of diabetes in plasma and urine samples46,47,48. It is inversely associated with several components of metabolic syndrome including obesity, hypertension, and hyperlipemia49. Mouse model studies suggest increased betaine metabolism in diabetes, which could be expected to lower plasma betaine50. In agreement with these findings, a reduction of betaine level is an important contribution to DM development.
A decreased level of L-carnitine and an increased level of dityrosine and 3-dehydroxycarnitine were detected in patients progressed to DM. These results are consistent with an interesting study investigating the relationship between the consumption of red meat and the development of associated diseases51. L-carnitine is known to be a long-chain fatty acid transporter in the “Mitocondrial L-carnitine Shuttle Pathway”52, and 3-dehydroxycarnitine has been identified as an intermediate metabolite in the intestinal bacterial catabolism of L-carnitine53, which may give a hint the involvement of gut microbiota in DM development. Dityrosine has been proposed as a biomarker of oxidative stress under a variety of conditions and biological systems including aging, exposure to oxygen free radicals, nitrogen dioxide, and lipid hydroperoxides54. Increases in dityrosine levels have been associated with pathologies such as atherosclerosis, Alzheimer’s Disease, and so on55,56. Together, these findings reflect impaired mitochondrial β-oxidation and perturbed fatty acid metabolism in the development of insulin resistance.
To make it better to apply the identified biomarkers for prediction of pre-DM prognosis in the long run in clinical setting, the key of this study was to select and combine several specific biomarkers for establishing a noninvasive and accurate predict method for prognosis of pre-DM. The candidate biomarker selection rationale was as follows: first, the biomarkers must be confirmed by standards; second, the biomarkers with high VIPs in the pattern recognition analysis and significant discrepancy between groups; and last, the biomarkers in the logistic regression equation with higher AUCs and predictive sensibility and specificity. As a result, five biomarkers (20-Hydroxy-leukotriene E4, Lysopc(20:4), 5-methoxytryptamine, Endomorphin-1, Lysopc(20:3)) and five biomarkers (Iso-valeraldehyde, linoleic acid, Lysopc(18:1), 2-Pyrroloylglycine, Dityrosine) were included in the predictive equation of NGR and DM, respectively.
In summary, our study comprehensively captured alterations in the human metabolome associated with different glucose tolerance outcomes of pre-DM in a longitudinal cohort study. A decreased glycerophosphoslipid metabolism and balanced oxidation and antioxidation contributed to the transition to NGR, while impaired amino acid metabolism and perturbed mitochondrial β-oxidation were associated with the development of DM. Targeting the pathways that involve in these newly prognosis biomarkers would be beneficial for the regression to NGR and the early prevention of DM among participants with pre-DM.
Subjects and Methods
Ethics Statement
This study was approved by Ruijin Hospital Ethics Committee (approval no. 2014–114).
Written informed consent signed by each of participants was provided before blood samples were taken. All methods were carried out in accordance with the relevant guidelines and regulations.
Study design and Subjects
This study was from a population-based prospective cohort study of 2132 men and women aged 18–76 years, from November 2002 to January 2003, among whom 778 participants were pre-DM at baseline. The follow-up visit was conducted from July 2013 to October 2014 and 526 participants who were pre-DM at baseline were followed, among whom 334 individuals both answered questionnaires and had plasma glucose measurement during an oral glucose tolerance test (OGTT). Serum lipid profile and liver function were also assayed. After excluded the individuals on anti-diabetes medication or with serious liver, renal dysfunction and cancer, the remaining 108 individuals were included in our final analysis. This study design has been described previously57,58,59.
According to different glycemic outcomes at follow up, the 108 participants were divided into 3 groups. 20 participants progressed to diabetes DM, 20 regressed to NGR, and 68 remained at pre-DM. Details of the study population are presented in Supplemental Fig. S3.
Venous blood samples were collected at baseline and follow-up. The glucose level was measured by means of glucose oxidase method. Pre-DM and diabetes were diagnosed according to American Diabetes Association (ADA) 2010 Guidelines60. Pre-DM refers to subjects with impaired fasting glucose (fasting glucose ranging from 5.6 to < 7.0 mmol/L, as well as 2-hour glucose < 7.8 mmol/L) and subjects with impaired glucose tolerance (2-hour glucose ranging from 7.8 to < 11.1 mmol/L). Both impaired fasting glucose and impaired glucose tolerance were confined to non-diabetic fasting and 2-hour concentrations. Fasting plasma samples were used to analyze biochemical indexes and metabolomics.
Sample collection and preparation
Fasting blood samples were drawn under sterile conditions from an antecubital vein of all the study participants between 6:30 and 9:30 after a 12-hour overnight fast, and were collected directly into heparinized tubes. The tubes were centrifuged at 12000 g for 10 min and the supernatant (plasma sample) was aspirated and stored at −80 °C until analysis.
The plasma sample (100 μl) was thawed at 4 °C. 100 μl of plasma was spiked with 300 μl mixed solution (methanol: acetonitrile = 3:2) and vigorously vortexed for 30 seconds. The sample solution was centrifuged at 12000 × g for 10 min at 4 °C, and the supernatant was analyzed using UPLC-QTOF-MS. Study samples were analyzed in random order using a random-number generator in Excel 2015 (Microsoft, Redmond, Washington). QC samples were prepared by mixing equal volumes (10 μl) of different individual plasma samples and one QC sample was run after every ten study sample injections throughout the analytical workflow.
UPLC-QTOF-MS conditions
In this study, a Waters ACQUITYTM ultra performance liquid chromatography system (Waters Corp., Milford, USA) coupled with aSynaptG2 quadrupole time-of-flight (Q/TOF) tandem mass spectrometer (Waters, Milford, MA) was used to perform the analysis of plasma samples. The set-up parameters for the UPLC-QTOF-MS analysis were as follows: A T3 C18 chromatographic column (Waters, 2.1 mm × 100 mm, 1.7 μm) was used to separate metabolites contained in plasma with column temperature set at 45 °C. The eluted solution was 0.1% formic acid combined with 5 mM ammonium acetate in water (A) and acetonitrile (B) with a flow rate of 300 μl/min. The gradient elution program for analysis of plasma samples was as follows: 0–1 min, A: 98%; 1–3 min, A: 98–50%; 3–8 min, A: 50–45%; 8–12 min, A: 45%; 12–17 min, A: 45%−10%; 17–20 min, A: 10–98%.
The MS parameters were set up as follows: the electrospray ionization source (ESI) interface operated with a positive mode, capillary voltage of 3000 V, sample cone voltage of 40 V, extraction cone voltage of 4.0 V, desolvation gas flow of 650 L/h at 450 °C, source temperature of 120 °C, and cone gas flow of 50 L/h. Centroid data were collected under a scan time of 0.25 s and an inter scan delay of 0.02 s condition in continuum mode which ranged from m/z 100 to m/z 1200 Da.
To avoid possible contamination and keep the signal stable, the Q-TOF mass spectrometer system was tuned for optimum accuracy and reproducibility using leucine-enkephalin (m/z 556.2771) as the lock mass in all analyses at a concentration of 0.5 μg/mL. The lock spray frequency was set at 5 s and the lock mass data were averaged over 10 scans. MSE was applied for the MS2 analysis with the low collision energy of 5 eV and the high collision energy of 30 eV.
Data extraction and multivariate statistics
The raw data produced by UPLC-QTOF-MS were initially processed using MarkerLynx Applications Manager version 4.1 (Waters Corp., Manchester, UK). The data were peak-detected and noise-reduced so that only true peaks are further processed by the software. The data were presented with the ion intensities corresponding the retention time and m/z for each peak. The main parameters were set as follows: retention time window 0.5–16.5 min, mass range 100–1200 Da, XIC window 0.02 min, automatically calculate peak width and peak-peak baseline noise, use the raw data during the deconvolution procedure, marker intensity threshold (count) 1000, mass window 0.02 Da, retention time windows 0.2 min, noise elimination level 6.0, and retain the isotopic peaks. The internal standard was used for data quality control and data normalization (reproducibility). The ion peaks generated by the internal standard were removed and the metabolites were filtered by the QC samples.
The 80% rule was applied to treat the missing values61 and a data matrix that consisted of the ion intensities corresponding the retention time and m/z for each peak was generated and then exported to Simca-P software (v13.0, Umetrics, Umea, Sweden) followed by a series of pattern recognition (PR) methods. Multivariate statistical analyses, including principal component analysis (PCA) and a partial least squares discriminant analysis (PLS-DA) were carried out using SIMCA-P 13.0 software. The score plots from PLS-DA showed the differentiation of metabolic profiles of different groups. In addition, loading plots indicated the variables contributing to the classification. The quality of the model was described by the cross-validation parameter Q2 (cum), and R2Y, which represents the total explained variation for the X matrix. After the analysis of the three groups, pair-wise analysis (NGR vs pre-DM, DM vs pre-DM and NGR vs DM) was conducted to searching for the discrepant metabolites between groups.
Identification of potential biomarkers and metabolic pathway analysis
The metabolites responsible for the separation of metabolic profiles of the pair-wise groups were obtained based on a variable importance in projection (VIP) threshold (VIP > 1 represented higher influence on the classification)17 from PLS-DA models accompanied with loading plots and a statistical test for difference (p < 0.05 was considered significant). A two-tailed t test or a nonparametric Mann-Whitney test was used for significance evaluation following data normality test with Shapiro-Wilk tests, which was performed with the R statistical software 3.3.2 for Windows.
A metabolite was detected and identified based on accurate mass, retention time, MS information and metabolite structure information from related databases: METLIN (https://metlin.scripps.edu/index.php) and HMDB (http://www.hmdb.ca/). Some of the metabolites were confirmed by comparison of retention time and fragmentation pattern with authentic standards.
The pathway analysis and network of potential biomarkers contributing to the classification between groups was carried out by IPA software (IPA, Ingenuitys Systems, http://www.ingenuity.com).
The schematic flow chart of the metabolic profiling and biomarker identification and optimization strategy used in the study is shown in Fig. 5.
Schematic flow chart of the metabolomics analysis in the study. NGR: normal glucose regulation; Pre-DM: pre-diabetes; DM: diabetes; UPLC-QTOF-MS: Ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry; VIP: variable importance in the projection; ROC: receiver operating characteristic curve; AUC: area under the curve.
Statistical analysis
Demographic and biochemical characteristics were presented as the mean (±SD) for continuous variables and % for categorical variables. Data normality and homogeneity of continuous variables were confirmed with Shapiro-Wilk tests and Levene tests, respectively. If data were normally distributed and variances were equal, data were analyzed by means of one-way analysis of variance (ANOVA) with LSD test; Otherwise, Kruskal-Wallis test with a nonparametric two-tailed Mann-Whitney test was used (alpha was adjusted to 0.05/3 here). And categorical variables were analyzed by χ2 test or Fisher’s exact test. Difference was considered statistically significant when p < 0.05.
Multiple logistic regression analysis of the potential metabolites was performed and receiver operating characteristics (ROC) analysis was used to evaluate predictive ability of potential metabolic biomarkers. Area under the curve (AUC), best cut-off point, sensitivity and specificity were determined using the maximum value of the Youden index. The analyses were performed using SPSS software version 23.0 (IBM Corp., USA).
Change history
21 November 2017
A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
References
Tabak, A. G., Herder, C., Rathmann, W., Brunner, E. J. & Kivimaki, M. Prediabetes: a high-risk state for diabetes development. Lancet (London, England) 379, 2279–2290, doi:10.1016/s0140-6736(12)60283-9 (2012).
Cefalu, W. T. et al. Update and Next Steps for Real-World Translation of Interventions for Type 2 Diabetes Prevention: Reflections From a Diabetes Care Editors’ Expert Forum. Diabetes care 39, 1186–1201, doi:10.2337/dc16-0873 (2016).
Xu, Y. et al. Prevalence and control of diabetes in Chinese adults. Jama 310, 948–959, doi:10.1001/jama.2013.168118 (2013).
Nathan, D. M. et al. Impaired fasting glucose and impaired glucose tolerance: implications for care. Diabetes care 30, 753–759, doi:10.2337/dc07-9920 (2007).
Phillips, L. S., Ratner, R. E., Buse, J. B. & Kahn, S. E. We can change the natural history of type 2 diabetes. Diabetes care 37, 2668–2676, doi:10.2337/dc14-0817 (2014).
Gerstein, H. C. et al. Effect of rosiglitazone on the frequency of diabetes in patients with impaired glucose tolerance or impaired fasting glucose: a randomised controlled trial. Lancet 368, 1096–1105, doi:10.1016/s0140-6736(06)69420-8 (2006).
Knowler, W. C. et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346, 393–403, doi:10.1056/NEJMoa012512 (2002).
Tuomilehto, J. et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med 344, 1343–1350, doi:10.1056/nejm200105033441801 (2001).
Association, A. D. Prevention or Delay of Type 2 Diabetes. Jan, S31–S32 (2015).
Guasch-Ferre, M. et al. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes care 39, 833–846, doi:10.2337/dc15-2251 (2016).
Pallares-Mendez, R., Aguilar-Salinas, C. A., Cruz-Bautista, I. & Del Bosque-Plata, L. Metabolomics in diabetes, a review. Annals of medicine 48, 89–102, doi:10.3109/07853890.2015.1137630 (2016).
Wang, T. J. et al. Metabolite profiles and the risk of developing diabetes. Nat Med 17, 448–453, doi:10.1038/nm.2307 (2011).
Zhao, X. et al. Metabonomic fingerprints of fasting plasma and spot urine reveal human pre-diabetic metabolic traits. Metabolomics 6, 362–374, doi:10.1007/s11306-010-0203-1 (2010).
Emwas, A.-H. M., Salek, R. M., Griffin, J. L. & Merzaban, J. NMR-based metabolomics in human disease diagnosis: applications, limitations, and recommendations. Metabolomics 9, 1048–1072, doi:10.1007/s11306-013-0524-y (2013).
Van, Q. N. & Veenstra, T. D. How close is the bench to the bedside? Metabolic profiling in cancer research. Genome medicine 1, 5, doi:10.1186/gm5 (2009).
Chen, T. et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Molecular & cellular proteomics: MCP 10, M110.004945, doi:10.1074/mcp.M110.004945 (2011).
Cheng, Y. et al. Distinct urinary metabolic profile of human colorectal cancer. Journal of proteome research 11, 1354–1363, doi:10.1021/pr201001a (2012).
Liu, T. et al. Comprehensive analysis of serum metabolites in gestational diabetes mellitus by UPLC/Q-TOF-MS. Analytical and bioanalytical chemistry 408, 1125–1135, doi:10.1007/s00216-015-9211-3 (2016).
Wang, M. et al. Biomarkers identified by urinary metabonomics for noninvasive diagnosis of nutritional rickets. Journal of proteome research 13, 4131–4142, doi:10.1021/pr500517u (2014).
Wu, Q. et al. Evidence for Chronic Kidney Disease-Mineral and Bone Disorder Associated With Metabolic Pathway Changes. Medicine 94, e1273, doi:10.1097/md.0000000000001273 (2015).
Zhang, Z. H. et al. Metabolomics insights into chronic kidney disease and modulatory effect of rhubarb against tubulointerstitial fibrosis. Scientific reports 5, 14472, doi:10.1038/srep14472 (2015).
Li, Y. et al. Screening, verification, and optimization of biomarkers for early prediction of cardiotoxicity based on metabolomics. Journal of proteome research 14, 2437–2445, doi:10.1021/pr501116c (2015).
Lundstedt, T. et al. Experimental design and optimization. Chemometrics and Intelligent Laboratory Systems 42, 3–40, doi:10.1016/S0169-7439(98)00065-3 (1998).
Stephan, C., Wesseling, S., Schink, T. & Jung, K. Comparison of eight computer programs for receiver-operating characteristic analysis. Clinical chemistry 49, 433–439 (2003).
Hsu, F. F. et al. Electrospray ionization mass spectrometric analyses of changes in tissue phospholipid molecular species during the evolution of hyperlipidemia and hyperglycemia in Zucker diabetic fatty rats. Lipids 35, 839–854 (2000).
Perassolo, M. S. et al. Fatty acid composition of serum lipid fractions in type 2 diabetic patients with microalbuminuria. Diabetes care 26, 613–618 (2003).
Berridge, M. J. Inositol trisphosphate and calcium signalling. Nature 361, 315–325, doi:10.1038/361315a0 (1993).
Exton, J. H. Phosphatidylcholine breakdown and signal transduction. Biochimica et biophysica acta 1212, 26–42 (1994).
Floegel, A. et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 62, 639–648, doi:10.2337/db12-0495 (2013).
Wang-Sattler, R. et al. Novel biomarkers for pre-diabetes identified by metabolomics. Molecular systems biology 8, 615, doi:10.1038/msb.2012.43 (2012).
Wang, T. J. et al. Metabolite Profiles and the Risk of Developing Diabetes. Nature medicine 17, 448–453, doi:10.1038/nm.2307 (2011).
Ma, H. & Patti, M. E. Bile acids, obesity, and the metabolic syndrome. Best practice & research. Clinical gastroenterology 28, 573–583, doi:10.1016/j.bpg.2014.07.004 (2014).
Sonoki, K. et al. Relations of lysophosphatidylcholine in low-density lipoprotein with serum lipoprotein-associated phospholipase A2, paraoxonase and homocysteine thiolactonase activities in patients with type 2 diabetes mellitus. Diabetes research and clinical practice 86, 117–123, doi:10.1016/j.diabres.2009.08.014 (2009).
Yea, K. et al. Lysophosphatidylcholine activates adipocyte glucose uptake and lowers blood glucose levels in murine models of diabetes. The Journal of biological chemistry 284, 33833–33840, doi:10.1074/jbc.M109.024869 (2009).
Tan, D. X. et al. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptamine: comparisons across species. Journal of pineal research 61, 27–40, doi:10.1111/jpi.12336 (2016).
Manchester, L. C. et al. Melatonin: an ancient molecule that makes oxygen metabolically tolerable. Journal of pineal research 59, 403–419, doi:10.1111/jpi.12267 (2015).
Zhang, H. M. & Zhang, Y. Melatonin: a well-documented antioxidant with conditional pro-oxidant actions. Journal of pineal research 57, 131–146, doi:10.1111/jpi.12162 (2014).
Poeggeler, B., Reiter, R. J., Tan, D. X., Chen, L. D. & Manchester, L. C. Melatonin, hydroxyl radical-mediated oxidative damage, and aging: a hypothesis. Journal of pineal research 14, 151–168 (1993).
Letra-Vilela, R. et al. Distinct roles of N-acetyl and 5-methoxy groups in the antiproliferative and neuroprotective effects of melatonin. Molecular and cellular endocrinology 434, 238–249, doi:10.1016/j.mce.2016.07.012 (2016).
Silva, R. O. et al. Phytol, a diterpene alcohol, inhibits the inflammatory response by reducing cytokine production and oxidative stress. Fundamental & clinical pharmacology 28, 455–464, doi:10.1111/fcp.12049 (2014).
Han, J., Hamilton, J. A., Kirkland, J. L., Corkey, B. E. & Guo, W. Medium-chain oil reduces fat mass and down-regulates expression of adipogenic genes in rats. Obesity research 11, 734–744, doi:10.1038/oby.2003.103 (2003).
Yu, D. et al. Plasma metabolomic profiles in association with type 2 diabetes risk and prevalence in Chinese adults. Metabolomics 12, doi:10.1007/s11306-015-0890-8 (2016).
Yan-Do, R. & MacDonald, P. E. Impaired “glycine”-mia in type 2 diabetes and potential mechanisms contributing to glucose homeostasis. Endocrinology, doi:10.1210/en.2017-00148 (2017).
Takashina, C. et al. Associations among the plasma amino acid profile, obesity, and glucose metabolism in Japanese adults with normal glucose tolerance. Nutrition & metabolism 13, 5, doi:10.1186/s12986-015-0059-5 (2016).
Wang, W. et al. Glycine metabolism in animals and humans: implications for nutrition and health. Amino acids 45, 463–477, doi:10.1007/s00726-013-1493-1 (2013).
Schartum-Hansen, H. et al. Assessment of urinary betaine as a marker of diabetes mellitus in cardiovascular patients. PloS one 8, e69454, doi:10.1371/journal.pone.0069454 (2013).
Svingen, G. F. et al. Prospective Associations of Systemic and Urinary Choline Metabolites with Incident Type 2 Diabetes. Clinical chemistry 62, 755–765, doi:10.1373/clinchem.2015.250761 (2016).
Walford, G. A. et al. Metabolite Profiles of Diabetes Incidence and Intervention Response in the Diabetes Prevention Program. Diabetes 65, 1424–1433, doi:10.2337/db15-1063 (2016).
Konstantinova, S. V. et al. Divergent associations of plasma choline and betaine with components of metabolic syndrome in middle age and elderly men and women. The Journal of nutrition 138, 914–920 (2008).
Ratnam, S. et al. Effects of diabetes and insulin on betaine-homocysteine S-methyltransferase expression in rat liver. American journal of physiology. Endocrinology and metabolism 290, E933–939, doi:10.1152/ajpendo.00498.2005 (2006).
Rombouts, C. et al. Untargeted metabolomics of colonic digests reveals kynurenine pathway metabolites, dityrosine and 3-dehydroxycarnitine as red versus white meat discriminating metabolites. Scientific reports 7, 42514, doi:10.1038/srep42514 (2017).
Dambrova, M. & Liepinsh, E. Risks and benefits of carnitine supplementation in diabetes. Experimental and clinical endocrinology & diabetes: official journal, German Society of Endocrinology [and] German Diabetes Association 123, 95–100, doi:10.1055/s-0034-1390481 (2015).
Koeth, R. A. et al. gamma-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell metabolism 20, 799–812, doi:10.1016/j.cmet.2014.10.006 (2014).
Giulivi, C., Traaseth, N. J. & Davies, K. J. Tyrosine oxidation products: analysis and biological relevance. Amino acids 25, 227–232, doi:10.1007/s00726-003-0013-0 (2003).
Al-Hilaly, Y. K. et al. A central role for dityrosine crosslinking of Amyloid-beta in Alzheimer’s disease. Acta neuropathologica communications 1, 83, doi:10.1186/2051-5960-1-83 (2013).
DiMarco, T. & Giulivi, C. Current analytical methods for the detection of dityrosine, a biomarker of oxidative stress, in biological samples. Mass spectrometry reviews 26, 108–120, doi:10.1002/mas.20109 (2007).
Song, X. et al. Gender-related affecting factors of prediabetes on its 10-year outcome. BMJ open diabetes research & care 4, e000169, doi:10.1136/bmjdrc-2015-000169 (2016).
Qiu, M. et al. Effects of prediabetes mellitus alone or plus hypertension on subsequent occurrence of cardiovascular disease and diabetes mellitus: longitudinal study. Hypertension. (Dallas, Tex.: 1979) 65, 525–530, doi:10.1161/hypertensionaha.114.04632 (2015).
Tian, J. Y. et al. Birth weight and risk of type 2 diabetes, abdominal obesity and hypertension among Chinese adults. Eur J Endocrinol 155, 601–607, doi:10.1530/eje.1.02265 (2006).
Diagnosis and classification of diabetes mellitus. Diabetes care 33 Suppl 1, S62–69, doi:10.2337/dc10-S062 (2010).
Dunn, W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nature protocols 6, 1060–1083, doi:10.1038/nprot.2011.335 (2011).
Acknowledgements
We are indebted to staff and participants of the study for their important contributions. We gratefully acknowledge grants supported by the National Natural Science Foundation of China (No. 81270935 and 81573581), Transform Medicine Innovation Foundation of Shanghai Jiao Tong University School of Medicine (15ZH2001), the fund of the Key Laboratory of Stem Cell Biology of Chinese Academy of Sciences (No. 201601), Shanghai Municipal Health Bureau Foundation (20114301), the Program of Shanghai Subject Chief Scientist (17XD1403500) and Shanghai Science and Technology Commission Project (15401902400).
Author information
Authors and Affiliations
Contributions
J.T. and L.Y. designed the study. H.Z., R.T., W.T., Q.Y., M.Q., A.X. performed experiments. H.Z. and R.T. analyzed data and wrote the paper. L.D., S.S., H.Z., J.T. and L.Y. reviewed and edited the manuscripts. All authors reviewed and approved the final article.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Change History: A correction to this article has been published and is linked from the HTML version of this paper. The error has been fixed in the paper.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A correction to this article is available online at https://doi.org/10.1038/s41598-017-14856-1.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zeng, H., Tong, R., Tong, W. et al. Metabolic Biomarkers for Prognostic Prediction of Pre-diabetes: results from a longitudinal cohort study. Sci Rep 7, 6575 (2017). https://doi.org/10.1038/s41598-017-06309-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-06309-6
This article is cited by
-
Identification of metabolic markers predictive of prediabetes in a Korean population
Scientific Reports (2020)
-
Pre-meal protein intake alters postprandial plasma metabolome in subjects with metabolic syndrome
European Journal of Nutrition (2020)
-
Increasing Acyl CoA thioesterase activity alters phospholipid profile without effect on insulin action in skeletal muscle of rats
Scientific Reports (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.