Abstract
The importance of microbial natural products has been widely demonstrated in the search for new antibiotics. However, the functional role of microbial metabolites in nature remains to be deciphered. Several natural products are known to mediate microbial interactions through metabolic exchange. One approach to investigate metabolic exchange in the laboratory is through microbial interactions. Here, we describe the chemical study of selected endophytes isolated from the Brazilian medicinal plant Lychnophora ericoides by pairwise inter-kingdom interactions in order to correlate the impact of co-cultivation to their metabolic profiles. Combining mass spectrometry tools and NMR analyses, a total of 29 compounds were identified. These compounds are members of polyene macrocycles, pyrroloindole alkaloids, angucyclines, and leupeptins chemical families. Two of the identified compounds correspond to a new fungal metabolite (29) and a new actinobacterial angucycline-derivative (23). Our results revealed a substantial arsenal of small molecules induced by microbial interactions, as we begin to unravel the complexity of microbial interactions associated with endophytic systems.
Similar content being viewed by others
Introduction
Endophytes, reported for the first time more than a century ago1, are described as microorganisms inhabiting plant tissues without causing pathogenic effects into their host2, 3. These microorganisms are part of complex systems of balanced interactions mediated by natural products between endophytes and their host4,5,6. The biotechnological interest in endophytic microbes due to their potential as sources of natural products began with the discovery of paclitaxel7, and subsequent reports of other important bioactive molecules from endophytes, such as deoxypodophyllotoxin8, maytansine9, 10, campthotecin11, 12, among many others13.
In the recent years, the investigation of natural products from microorganisms by using co-culture strategies has increased14,15,16. The co-culture approach is one of the several other strategies to induce the expression of cryptic microbial metabolites17, and increase metabolic diversity15, 18, 19. Since microorganisms are part of natural consortia6, the discovery of natural products from microbial interactions, not present in axenic cultures, is expected. Studies have demonstrated the elicitation of microbial metabolites as a consequence of interactions from endophytes20, 21, including overproduction of paclitaxel22. Following this rational, it is a promising endeavor to study the chemistry of interacting microorganisms to reveal molecules that are instrumental for the establishment of complex communities23. Studies into the chemistry of endophytes, combined with investigation of their involvement in microbial interactions, would contribute to our understanding of their role in natural environments24, 25.
The co-culture strategy, combined with mass spectrometry (MS) approaches, such as molecular networking26,27,28, and automatic search against experimental and in silico databases29, can lead to the detection and identification of microbial metabolites. MS-based molecular networking uses tandem MS (MS/MS) to correlate chemical entities by structural similarity, identify known molecules and accelerate the discovery of new ones27, 30. However, the challenge remains in the correct identification of most of these chemical entities31. Therefore, the fully characterization by Nuclear Magnetic Resonance (NMR) and MS of purified compounds is pivotal to confirm chemical structures27, 30.
Several studies investigating natural products from the Brazilian medicinal plant Lychnophora ericoides have been published32,33,34,35,36,37,38,39,40,41,42,43. Due to the importance of this plant in folk medicine42, 43, and the interesting biosynthetic potential of endophytic microorganisms13, we initiated the investigation of natural products from endophytes of L. ericoides 44. A total of sixteen compounds were isolated from endophytic actinobacteria of L. ericoides, revealing these microorganisms as a promising source of natural products44. This potential was also demonstrated in a recent study where we investigated the chemical arsenal produced by one of these endophytic actinobacteria strains, Streptomyces albospinus RLe745. A total of seven compounds were isolated, including three of them that were previously unknown as natural products45. Although S. albospinus RLe7 showed a low cytotoxicity against cancer cell lines44, it induced a particular fungal response in the endophytic Coniochaeta sp. FLe4 due to the production of amphotericin B (1)45. These results suggest endophytes may have the potential to produce chemical compounds during microbial interactions.
With the purpose of investigating the chemical arsenal of endophytic microorganisms, randomly selected strains from our collection of endophytic strains of the Brazilian medicinal plant L. ericoides were submitted to pairwise microbial interactions. In this study, we present the microbial interactions between five actinobacteria (S. cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7, Streptomyces sp. RLe9 and Kytasatospora cystarginea RLe10) and two fungi (Colletotrichum boninense FLe8.1 and Coniochaeta sp. FLe4). Particularly, chemical families from S. mobaraensis RLe3, which showed a high cytotoxicity activity against cancer cells in our previous study44, and S. albospinus RLe7, an amphotericin B (1) producer45, were identified by performing complementary approaches involving mono- and co-cultures, characterization of isolated compounds via NMR and MS, as well as molecular networking workflow.
Therefore, we report the identification of chemical families (pyrroloindole alkaloids, polyene macrocycles, leupeptins and angucyclines) produced by endophytic actinobacteria, characterization of cytotoxic agents (angucycline-derivatives) from S. mobaraensis RLe3, inducer activity of amphotericin B (1) (polyene macrocycle) from S. albospinus RLe7 by isolating a new fungal compound (29) from Coniochaeta sp. FLe4, characterization of a new compound from S. mobaraensis RLe3 (23) (angucycline-derivative), and showed the impact of inter-kingdom interactions between actinobacteria and fungi. Finally, we contribute with the annotation of the experimental MS/MS spectra from the 29 identified microbial natural products to the GNPS library (http://gnps.ucsd.edu)27.
Results and Discussion
Pairwise inter-kingdom interactions
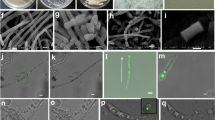
Microbial interactions were performed between the endophytic strains of actinobacteria (S. cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7, Streptomyces sp. RLe9 and Kytasatospora cystarginea RLe10) and fungi (Colletotrichum boninense FLe8.1 and Coniochaeta sp. FLe4) (Supplementary Fig. S1). It was observed that co-culturing the fungi Colletotrichum boninense FLe8.1 and Coniochaeta sp. FLe4 with actinobacteria had different impact on fungal development (Fig. 1). The actinobacteria affected fungal growth of the closest colonies of the fungus Coniochaeta sp. FLe4, but did not completely inhibit the fungus (Fig. 1a–e). A red pigmented phenotype was visible when Coniochaeta sp. FLe4 was co-cultured with four of the actinobacteria (Fig. 1a–c and e). This phenotype was most pronounced during the interaction with S. albospinus RLe7 (Fig. 1c). The colony of C. boninense FLe8.1 most proximal to actinobacteria did not grow in any of the interactions and no pigmentation was observed (Fig. 1g–k). These observations suggested the presence of antifungal compounds produced by actinobacteria.
Pairwise inter-kingdom interactions involving endophytic fungi Coniochaeta sp. FLe4 and C. boninense FLe8.1 with endophytic actinobacteria from L. ericoides. Actinobacteria: S. cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7, Streptomyces sp. RLe9, K. cystarginea RLe10. Fungi: Coniochaeta sp. FLe4 and C. boninense FLe8.1. Every interaction involved two colonies of each microorganism. Photos taken after 96 hours of cultivation.
Identification of antibiotics/antifungals from actinobacteria by molecular networking
To reveal the small molecules produced during the pairwise inter-kingdom interactions, particularly the metabolite responsible for the induction of the red phenotype of Coniochaeta sp. FLe4 (Fig. 1c), molecular networking workflow was performed. Molecular network of microbial interactions enabled easy visualization of specific microbial metabolic features and identification of the producer microorganisms (Supplementary Fig. S2). Briefly, in a molecular network, every node represents one chemical entity, while clusters of nodes correspond to structurally related molecules based on similarity of their MS/MS spectra pattern27. While some nodes are annotated as previously characterized compounds, clusters including those metabolites are called molecular families46. A complementary strategy composed by the built-in automatic library search by GNPS27, manual confirmation based on MS/MS fragmentation pattern, accurate mass and NMR data of isolated compounds were used to verify and confirm the molecules and chemical classes described as follows.
One of the metabolites previously identified from S. albospinus RLe7 corresponds to the pyrroloindole alkaloid physostigmine (compound 2)44. As it is a characterized natural product from this endophytic actinobacterium, the detection of physostigmine during microbial interaction was expected. In addition, the MS/MS fragmentation pathway of this well-known anticholinesterase compound has been reported47. The node corresponding to physostigmine clustered with other potentially related molecules (Figs 2 and S3–5). Two of them where consistent with the chemical formula of antibiotics TAN1169A (compound 3) and B (compound 4), previously reported natural products48, recently found to be intermediates of the physostigmine biosynthesis49. Although physostigmine was consistently detected from samples of mono- and co-cultures of S. albospinus RLe7, its biosynthesis was negatively affected by microbial interactions (Supplementary Fig. S6). Compounds 3 and 4 were also detected in mono- and co-culture samples (Supplementary Figs S7 and S8), and represented as yellow nodes (Fig. 2). Therefore, these three pyrroloindole alkaloid analogues were detected in mono- and co-culture samples. Physostigmine (2) has no reported antifungal activity, and it was not responsible for the induction of the red phenotype of Coniochaeta sp. FLe4 when interacting with S. albospinus RLe745. However, the insecticidal activity of this compound against silkworm larvae and the importance of the N-8 methyl group in this bioactivity was suggested50. It is possible that physostigmine, as well as its analogues (compounds 3 and 4), play an ecological role by protecting plants against insects, illustrating the importance of endophytes for symbiotic interactions.
Pyrroloindole alkaloids-cluster from the molecular network of interactions among endophytic microorganisms from L. ericoides. Nodes from co-cultures are red, nodes from several strains (non-specified microbial origin) are purple, nodes observed in mono- and co-cultures involving S. albospinus RLe7 are in yellow.
The automatic library search by GNPS suggested the presence of amphotericin-derivatives by similarity of fragmentation patterns. However, manual verification of MS/MS and accurate mass were necessary to confirm the presence of amphotericin B (compound 1) (Figs 3 and S9). MS/MS fragmentation of this class of compounds under our acquisition parameters was useful for identification of related compounds, due to their characteristic fragmentation patterns51. Therefore, it was confirmed that S. albospinus RLe7 also produced macrocyclic polyenes, related to amphotericin B (compound 1). This cluster suggested the presence of several analogues, such as amphotericin A (compound 5) (Supplementary Fig. S10), in consistency with the previous reports about coproduction of amphotericin A (5) and B (1)52, 53. Besides that, some “impurities”54, or even sub-products that may correspond to extracting adducts, such as amphotericin X or B2 (compound 6) (Supplementary Fig. S11), which possess a methoxy group at C-13 position55, were identified. Additional putative analogues with mass differences were consistent with variations in the oxygenation and double bond patterns, such as the ions of m/z 888 (compound 7), m/z 906 (compound 8), m/z 940 (compound 9), m/z 942 (compound 10) and m/z 960 (compound 11). This cluster was also interesting because of the presence of some analogues with molecular formulas of compounds obtained by genetically engineered strains, such as 8-deoxyamphotericin A (compound 12) (Supplementary Fig. S12) and B (compound 13) (Supplementary Fig. S13)56. Manual verification of the detected amphotericin-analogues enabled us to confirm that compounds 1, 5–13 were detected in mono- and co-cultures (Supplementary Figs S14–S23). The detection of the ions of m/z 888 and m/z 906 at low levels from samples of mono-cultures from S. albospinus RLe7 affected the acquisition of MS/MS, decreasing the quality of the spectra due to contribution of noise signals, resulting in both nodes represented as features only observed from co-cultures (red nodes, Fig. 3). Although some amphotericin analogues have been investigated from a pharmaceutical point of view57, their role in the environment remain to be characterized. Since we demonstrated the involvement of this chemical class in the induction of the described particular fungal response of Coniochaeta sp. FLe4, visualized as a red-pigmented phenotype45, chemical signaling is perhaps a potential role polyene macrocycles may have in nature.
Amphotericin-cluster from the molecular network of interactions among endophytic microorganisms from L. ericoides. Nodes from co-cultures are red, nodes observed only in mono-cultures are differentiated by a diamond shape, nodes originated from S. albospinus RLe7 in mono- and co-cultures are yellow and round nodes.
A widely distributed chemical class identified in this study corresponds to leupeptins (Fig. 4). The identification of leupeptin, m/z 427 (compound 14), based on the automatic library search by GNPS and manual verification of its fragmentation pattern (Supplementary Fig. S23), enabled the putative annotation of the ions of m/z 413 and m/z 505 as N-acetylleucylvalylargininal (compound 15) (or N-acetylvalylleucylargininal, compound 16) (Supplementary Fig. S25) and strepin P1 (17) (Supplementary Fig. S26), respectively. Additionally, the ion of m/z 441 was correctly identified as Leupeptin Pr-LL (compound 18) (Supplementary Fig. S27). Manual verification enabled us to confirm that leupeptins 14 and 15 (or 16) were produced by S. albospinus RLe7 and S. cattleya RLe1 (Supplementary Figs S28–S31). Strepin P1 (17) was produced by S. cattleya RLe1 (Supplementary Fig. S32) but also produced by K. cystarginea RLe10 (Supplementary Fig. S33). However, compound 17 was detected in mono-cultures involving K. cystarginea RLe10 and co-cultures with S. mobaraensis RLe3 (Supplementary Fig. S33), suggesting the other interactions led to inhibit its production. Finally, compound 18 was produced by S. cattleya RLe1 and detected in mono- and co-cultures (Supplementary Fig. S34). Protein inhibitors, such as leupeptin (14) and analogues (15–18), have been reported from Streptomyces 58,59,60 and also from marine Alteromonas 61. Compound 15, also proteinase inhibitor, previously isolated from Streptomyces and Alteromonas species, has been also obtained by synthesis61, 62, while the thrombin inhibitor 16, has been reported from S. flavogriseus 63, 64. Strepin P1 (17) was previously isolated from S. tanabaensis by bioactivity-guided fractionation showing proteinase inhibition65. Then, leupeptins are widely produced by actinomycetes59, which is also consistent with the obtained cluster of leupeptins visualized in the molecular network (Fig. 4), where leupeptin were not restricted to just one strain. Although the specific role of leupeptins in actinobacteria is not known, the involvement of leupeptins in morphological differentiation of mycelia has been demonstrated, giving a first insight into the natural role of this family of compounds during colony development of actinobacteria in the environment66, 67.
Leupeptin-cluster from the molecular network of interactions among endophytic microorganisms from L. ericoides. Nodes from co-cultures are red, nodes from several strains (unspecified microbial origin) are purple, nodes from mono- and co-cultures of S. cattleya RLe1 are in blue, nodes from mono- and co-cultures of S. mobaraensis RLe3 are in aquamarine, nodes from mono- and co-cultures of S. albospinus RLe7 are in yellow.
Angucycline-derivatives from S. mobaraensis RLe3
Since S. mobaraensis RLe3 also induced a red pigmented phenotype during co-culture in Coniochaeta sp. FLe4, we hypothesized that S. mobaraensis RLe3 also produced a compound responsible for eliciting this phenotype. Additional cultures in liquid and parboiled rice media of S. mobaraensis RLe3 were performed in order to investigate the chemical profile of this strain. A preliminary screening of inter-kingdom interactions in liquid media showed microbial interactions between S. mobaraensis RLe3 and Coniochaeta sp. FLe4 as an interesting co-culture for induction of microbial metabolites. Therefore, large-scale co-culture was performed with this interaction (Fig. 5). Chemical investigation of co-culture between S. mobaraensis RLe3 and Coniochaeta sp. FLe4 led to the isolation of three known compounds (Supplementary Fig. S35): aquayamycin (19) (Supplementary Figs S36–S40 and Supplementary Table S1), urdamycinone B (20) (Supplementary Figs S41–S45 and Supplementary Table S2) and galtamycinone (21) (Supplementary Figs S46–S50 and Supplementary Table S3). Overproduction of metabolites was observed when Coniochaeta sp. FLe4 was present during microbial interaction in liquid culture (Fig. 5). This observation was more obvious for urdamycinone B (20), but was also evident for aquayamycin (compound 19).
HPLC-DAD profile of extracts from microbial interaction between S. mobaraensis RLe3 and Coniochaeta sp. FLe4 in liquid culture. Overlaid chromatograms at 225 nm from culture media (black), S. mobaraensis RLe3 (blue), Coniochaeta sp. FLe4 (green), co-culture between RLe3 and FLe4 (red). Chemical structures from angucycline-derivatives isolated in this study. Urdamycinone B (20) is a representative metabolite overproduced by S. mobaraensis RLe3 during co-culture with Coniochaeta sp. FLe4 in liquid media.
Additionally, large-scale mono-culture on parboiled rice led to the isolation of two compounds (Supplementary Fig. S51): dehydroxyaquayamycin (22) (Supplementary Figs S52–S56 and Supplementary Table S4) and marangucycline A2 (23) (Supplementary Figs S57–S62 and Supplementary Table S5). The compound 23 showed similar NMR data and chemical formula consistent with a recently isolated new metabolite, marangucycline A68. Interestingly, NMR data acquired from this compound (Supplementary Figs S57–S62 and Supplementary Table S5) suggested the presence of a distinct sugar subunit. While marangucycline A contains a β-D-olivose and α-L-amicetose moiety, our data indicated a different configuration of the hydroxyl group at the C-4″ position, consistent with the presence of α–L-rhodinose instead of α-L-amicetose69. We named this analogue as marangucycline A2 (23). Additionally, the levels of compounds 22 and 23 were increased during microbial interaction in liquid co-culture of S. mobaraensis RLe3 and Coniochaeta sp. FLe4, as shown in Fig. 5.
Therefore, S. mobaraensis RLe3 was confirmed as an angucycline-producer by NMR and HR-MS. Although the mass fragmentation of this class of compounds has not been studied in detail, clusters representing angucycline-derivatives were visualized in the molecular network from microbial interactions in solid media (Fig. 6). Aquayamycin (19) (Supplementary Fig. S63), previously isolated from actinobacteria70,71,72, was also detected from mono- and co-cultures involving S. mobaraensis RLe3 (Supplementary Fig. S64). Additional angucycline-related compounds were also detected. For instance, the antibiotics DQ112A (compound 24)73 (Supplementary Figs S65 and S66) and BA12100MY1 (compound 25)74, (Supplementary Figs S67 and S68) detected in mono- and co-cultures (m/z 713 and m/z 469, respectively), clustered with aquayamycin (19) highlighting structural similarities (Fig. 6). Additionally, the ions of m/z 601 (compound 26), m/z 715 (compound 27) and m/z 711 (compound 28) were putatively annotated as angucycline analogues since their suggested chemical formula were consistent with at least two possible matches for each of them from the DNP database (Dictionary of Natural Products)75, and were detected in mono- and co-cultures involving S. mobaraensis RLe3 (Supplementary Figs S69-71). Urdamycinone B (20) was detected in mono- and co-cultures during interactions in solid media (Supplementary Figs S72 and S73). Urdamycinone B (20) was previously reported as a member of angucyclinones with an anthraquinone chromophore76. Galtamycinone (21)77, 78, which has been already reported from microbial sources79, was detected in mono- and in co-cultures and its fragmentation pattern showed similarity to urdamycinone B (20) (Supplementary Figs S74 and S75), resulting in both of them clustered together (Fig. 6). Dehydroxyaquayamycin (22), previously described as natural product from marine actinobacteria80, originally obtained as dehydrated product of aquayamycin (19)72, was detected in mono- and co-cultures involving S. mobaraensis RLe3 (Supplementary Figs S76 and S77). Dehydroxyaquayamycin (22) was found in low abundance which resulted in fragment spectra with additional peaks from chemical noise. Consequently, a separate two-nodes cluster was created by the algorithm (Fig. 6). The compound corresponding to marangucycline A2 (23) was detected in mono- and co-cultures of S. mobaraensis RLe3 (Supplementary Figs S78 and S79). Due to the detection of the corresponding ion of m/z 549 at low abundance, this compound (23) was represented as a single node in the molecular network (Fig. 6).
Angucyclines constitute the largest group of polycyclic aromatic polyketides with a wide range of biological activities81. Although this class of compounds has been widely reported, few studies have been carried out in order to reveal their function in nature. Recently, it was demonstrated the role of a “pseudo” gamma-butyrolactone receptor, which responded to exogenous angucyclines, in the regulation of the biosynthesis of endogenous antibiotics as well as its involvement in morphological development of Streptomyces 82. That study opened the door for understanding the ecological impact of natural products as mediators of microbial signaling. Although none of the angucycline-derivatives isolated in this study induced the red pigmentation in Coniochaeta sp. FLe4 (Supplementary Fig. S80), other compounds yet to be identified may be involved in this fungal response. On the other hand, the presence of angucycline-derivatives explained the high cytotoxic activities previously obtained from extracts of S. mobaraensis RLe344.
Structural elucidation of a new fungal metabolite
Based on the observation that the strongest red pigmented phenotype of the fungus Coniochaeta sp. FLe4 was induced when interacting with S. albospinus RLe7 (Fig. 1c), the identification of amphotericin B (1) from this actinobacteria, and the previous finding that this antifungal compound (1) is at least one of the responsible agents for inducing the red pigmented phenotype45, large-scale cultivation of Coniochaeta sp. FLe4 in presence of amphotericin B (1) was performed (Fig. 7). After several efforts to isolate the apparently induced compounds from the red pigment mixture, this fungal culture led to the isolation of compound 29 (Supplementary Figs S81 and S82), corresponding to the ion of m/z 265 (Fig. 8). Structural elucidation based on 1D and 2D NMR experiments (1H, gCOSY, gHSQC, gHMBC, NOESY, NOE DIFF, TOCSY) (Supplementary Figs S83–S90 and Supplementary Table S6) together with HR-MS-MS/MS led to a suggested molecular formula of C15H20O4, which was consistent with six degrees of unsaturation. Three methyl groups were detected as two duplets at δH 1.05 (J = 6.7 Hz) and 1.12 (J = 6.3 Hz) and one as a singlet at δH 1.87. HMBC correlation between hydrogens of a methoxy group at δH 3.86 to a carbon at δC 173.7 suggested the attachment of the methoxy group to a deshielded C sp2 which is linked to an oxygen atom. Two olefinic hydrogens at δH 6.19 (J = 15.7 Hz) and δH 7.11 (J = 15.7 Hz) showed trans correlation according to their J value. Experiment of NOE differential showed spatial correlation of the olefinic hydrogen at δH 5.57 to the methoxy group. Besides that, HMBC showed correlations of δH 5.57 to δC 101.4, δC 173.7 and δC 166.5. Additionally, HMBC correlations of δH 6.19 to δC 166.5 enabled to consistently suggest a free carboxyl group at δC 166.5 and the methoxy group to be linked to δC 173.7. Due to the low amount of sample it was not possible to directly detect the δC 160.7, which was attributed due to HMBC correlations to δH 6.14 and δH 6.19. The fragmentation pattern of compound 29 supports the possibility for a free carboxyl group that can lead to the product ion of m/z 220, while a subsequent loss of the methoxy group is consistent with the presence of the product ion of m/z 205. Together, our data support the proposal of the fungal compound 29 (Fig. 8).
Comparison of cultures involving Coniochaeta sp. FLe4 in presence and absence of amphotericin B. Coniochaeta sp. FLe4 cultured in amphotericin B-enriched ISP-2 medium at 2 µM (left) and in absence of amphotericin B, cultured only in ISP-2 medium (right). Photos were taken after six days of culture. For experimental details, see description for isolation of compound 29 in Materials and Methods section.
Node and representative MS/MS spectrum of m/z 265 (compound 29) from Coniochaeta sp. FLe4. Red node corresponds to detected metabolites from microbial interactions (co-cultures). Specifically, the node of m/z 265 was detected from samples of co-cultures involving Coniochaeta sp. FLe4 with S. cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7 and K. cystarginea RLe10.
In order to verify if induction of compound 29 was a consequence of microbial interaction, manual inspection of the raw LC-MS/MS data used to create the molecular network was performed. It was observed that the ion of m/z 265, corresponding to compound 29, was detected from microbial interactions involving Coniochaeta sp. FLe4 with the actinobacteria S. cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7 and K. cystarginea RLe10. However, it was not observed, at least at detectable levels, from the interaction with Streptomyces sp. RLe9 or any of the microbial mono-cultures (Supplementary Fig. S91). In addition, manual verification of the LC-MS/MS data from cultures of Coniochaeta sp. FLe4 in presence and absence of amphotericin B enabled to confirm the presence of compound 29 from cultures in amphotericin B-enriched medium (Supplementary Fig. S92).
Finally, our results showed that amphotericin B acts as an inducer of a fungal response leading to the production of the new fungal compound (29). As it was recently demonstrated, antibiotics such as trimethoprim, can induce the production of encrypted metabolites83. Our data showed that compound 29 was produced when Coniochaeta sp. FLe4 interacted with S. cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7 and K. cystarginea RLe10 but not with Streptomyces sp. RLe9 (Supplementary Fig. S91). This suggests a correlation with the observed red pigmented phenotype (Fig. 1a–c and e), not observed during interaction with Streptomyces sp. FLe9 (Fig. 1d). Notably, the fragmentation pattern of this compound shared no significant similarity with other nodes of the molecular network resulting in a single node or self-loop (Figs 8 and S2). This is one of the useful means to prioritize leads which correspond to structural uniqueness, as it has been illustrated for a Nocardiopsis metabolite, ciromicin A84. Further work focused on revealing the biological role of the fungal compound (29) will be necessary. Additionally, several induced metabolites may be part of the induced red pigmented complex since compound 29 is not a red pigment. Further investigation of this interesting finding may lead to the identification of other microbial natural products induced by compound 1.
Demonstrating the biological role for every molecule in nature is challenging. Therefore, the investigation of natural products in microbial communities is more rational since microorganisms are not alone in nature. Here we demonstrated the impact of microbial interactions among endophytes from L. ericoides that led to an increased production of some metabolites. We revealed the presence of an anticholinesterase compound (physostigmine-analogues), an antifungal compound (amphotericin-analogues), protease inhibitors (leupeptin-analogues) and cytotoxic (angucycline-analogues) chemical entities during investigation of microbial interactions (Summary Table 1). We demonstrated that metabolic exchange may induce chemical and phenotypical responses in inter-kingdom interactions among endophytic actinobacteria and fungi from L. ericoides. This study encourages us for further work to completely reveal both the chemistry and the biological role of small molecules from interacting endophytic microorganisms.
Materials and Methods
Strains and culture conditions
Endophytic strains
Endophytic microorganisms belong to the collection of the Laboratory of Chemistry of Microorganisms of the School of Pharmaceutical Sciences of Ribeirão Preto, University of São Paulo. The selected actinobacteria strains included in this work (Streptomyces cattleya RLe1, S. mobaraensis RLe3, S. albospinus RLe7, Streptomyces sp. RLe9 and Kytasatospora cystarginea RLe10) were isolated from the roots of L. ericoides and identified as previously described44. The fungus Coniochaeta sp. FLe4 was isolated from the leaves of the same plant and identified as described44, 45. The fungal strain Colletotrichum boninense FLe8.1 was isolated from the leaves of the same plant and its identification will be published soon. Permission of accessing and research with endophytic microorganisms from L. ericoides was provided by the Brazilian government under process number CNPq 010858/2014-8.
Reactivation of endophytic strains
Endophytic microorganisms were reactivated from their sterile mineral oil stocks44 in SFM agar medium (2 g soy flour (Sigma-Aldrich), 2 g agar (Sigma-Aldrich), 2 g mannitol (Sigma-Aldrich) per 100 mL of deionized water) for 7–10 days at 30 °C. Small pieces of SFM media-containing mycelia (5 mm diameter) were transferred to 4 mL of ISP-2 liquid (4 g yeast extract (Sigma-Aldrich), 10 g malt extract (Sigma-Aldrich), 4 g dextrose (Sigma-Aldrich) per 1 L of deionized water) and incubated for 72 hours in rotatory shaker at 200 rpm and 27–30 °C, obtaining a pre-culture.
Microbial interactions -inter-kingdom interactions- in solid media
In order to perform the microbial interactions, 1 μL of the pre-culture was spotted on Petri dishes (10 mm) containing 10 mL of ISP-2 agar media (15 g agar per 1 L of ISP-2). Mono-cultures of each microorganisms were performed by adding two 1 μL-spots of the pre-culture at 5 mm distance on the culture media. Co-cultures were performed following the same procedure, adding two 1 μL-spots of one microorganism pre-culture at 5 mm distance, and two 1 μL-spots of the other microorganism pre-culture at 5 mm distance (Supplementary Fig. S1 and Fig. 1). Four replicated samples from each mono- and co-culture were prepared. Microbial cultures were incubated at 30 °C during 96 h and then extraction was performed to prepare samples for subsequent LC- MS/MS analysis.
Large-scale of Streptomyces mobaraensis RLe3 in liquid co-culture with Coniochaeta sp. FLe4
Co-culture of S. mobaraensis RLe3 against Coniochaeta sp. FLe4 was performed as follows: 4 mL of the pre-culture in ISP-2 liquid containing S. mobaraensis RLe3, as described in the Strains and culture conditions section, and 12 plugs (5 mm diameter) of the reactivated Coniochaeta sp. FLe4 on SFM media containing fungal mycelia, as described in the Strains and culture conditions section, were transferred to 200 mL of ISP-2 in 500 mL-Erlenmeyer flasks. A total of 18 Erlenmeyer flasks were incubated on rotatory shaker at 30 °C and 200 rpm during 11 days.
Large-scale of Streptomyces mobaraensis RLe3 in mono-culture on parboiled rice
Mono-culture of Streptomyces mobaraensis RLe3 in parboiled rice was performed as described previously44, with some modifications briefly explained as following: 4 mL of the pre-culture in ISP-2 containing S. mobaraensis RLe3, as described in the Strains and culture conditions section, were transferred to 40 g of sterile parboiled rice contained in 500 mL-Erlenmeyer flasks. A total of 20 Erlenmeyer flasks were incubated at 30 °C during 40 days.
Large-scale of Coniochaeta sp. FLe4 in presence of amphotericin B
Amphotericin B (A2411 Sigma-Aldrich) was firstly dissolved in DMSO/MeOH 1:1 and added into sterilized ISP-2 agar at a final concentration of 2 µM. One hundred Petri dishes containing 10 mL of ISP-2 agar at a final concentration of 2 µM amphotericin B were prepared and inoculated with 500 µL of Coniochaeta sp. FLe4 from a 72 h pre-culture in ISP-2 liquid, as described in the Strains and culture conditions section. Plates were incubated under 30 °C for six days.
Instrumentation for purification via HPLC and NMR acquisition
HPLC purification of described compounds were carried out on a Shimadzu Prominence Nexera XR HPLC coupled to LC-6AD pumps, SPD-M20A Prominence Diode Array Detector, CTO-20A Prominence Column Oven, CBM-20A Communication Bus Module, and Class VP software. 1D and 2D NMR spectra were acquired on Bruker (R) – DRX500-Ultra Shield (R) (1H: 500.13 MHz, 13C: 125.77 MHz) and Bruker Avance III HD 600-MHz spectrometers.
Extraction, fractionation and purification procedures
Large-scale of Streptomyces mobaraensis RLe3 in liquid co-culture with Coniochaeta sp. FLe4
Extraction of the large-scale culture of the microbial interaction between S. mobaraensis RLe3 and Coniochaeta sp. FLe4 in liquid culture was carried as follows: after 11 days of co-culture, microbial cells were removed by filtration and the supernatant was extracted using SPE C-18. Briefly, SPE C-18 (10 g) cartridges (Phenomenex®) were conditioned with water, the filtrate was added to the cartridge and washed with water. The extract was eluted with MeOH and dried under vacuum. Sephadex LH20 (2.5 cm × 64 cm) was used for separation of ~975 mg of extract followed by further SPE C-18 fractionation before HPLC purification yielded the corresponding fractions containing the angucycline-related compounds 19, 20 and 21, respectively (Purification workflow and purified HPLC-DAD peaks: Supplementary Figs S35, S36, S41 and S46).
Large-scale of Streptomyces mobaraensis RLe3 in mono-culture on parboiled rice
Extraction of the large-scale culture for the scale up culture of S. mobaraensis RLe3 on parboiled rice was performed as follows: MeOH was added directly into the Erlenmeyer flasks containing the cultured microorganism, sonicated for 15 min, the methanolic extract was filtered and the elutant dried on a rotatory evaporator. Then, 1 g of the methanolic extract was subjected to fractionation by using Amberlite® XAD-16N resin (20–60 mesh, 4.5 cm × 32 cm column) with sequential elution using MeOH/water 1:1 until 100% MeOH was reached, followed by elution with 100% acetone. The acetone extract (202 mg) was subjected to further chromatographic steps by using Sephadex® LH20 (2.5 cm × 64 cm column). Additional purification steps included either SPE C-18 or Si (10 g cartridge), as well as HPLC yielded the corresponding fractions containing the angucycline-related compounds 22 and 23, respectively (Purification workflow purified HPLC-DAD peaks: Supplementary Figs S51, S52 and S57).
Purification of angucycline-derivatives
Further purification of the angucycline-derivatives was performed on a Phenomenex® Gemini C6-Phenyl 110 A semipreparative (5 µm × 250 mm × 10 mm) column. Solvent B: ACN, solvent A: water. Solvents were HPLC grade. The gradient employed for chromatographic separation was: 0–2 min 5% solvent B, 2–32 min linear gradient from 5% B to 100% B, 32–36 min 100% B, 36–40 min 100% to 5% B, 40–45 min 5% B at a flow rate of 4.7 mL/min throughout the run. Peaks of interest eluted at: 14.5 min (compound 19, 700 µg), 16.5 min (compound 20, 6.8 mg), 23.8 min (compound 21, 200 µg), 25.4 min (compound 22, 400 µg), 28.9 min (compound 23, 280 µg).
Large-scale of Coniochaeta sp. FLe4 in presence of amphotericin B
The large-scale cultures of Coniochaeta sp. FLe4 on ISP-2 media, enriched with amphotericin B (A2411 Sigma-Aldrich) at a final concentration of 2 µM, were extracted with 1 L of MeOH after six days of culture, sonicated for 15 min, filtered and dried by rotatory evaporation. The extract (1 g) was dissolved in 10 mL of milli-Q water and fractionated by using solid Phase Extraction (SPE) C-18 cartridge (10 g, Phenomenex®). The SPE cartridge was previously washed with MeOH and equilibrated with water. Gradient fractionation was carried out with water and increasing proportions of MeOH by 10%, collecting volumes of 100 mL until MeOH 100%. The eluted fraction of 60% MeOH contained the target compound (29). Further purification was carried out on a Phenomenex® Gemini C18 110 A semipreparative (5 µm × 250 mm × 10 mm) column under isocratic condition at 35% B. at a flow rate of 4.7 mL/min. Solvent B: ACN, solvent A: water, both solvents were HPLC grade. Peak of interest eluted at 15.9 min yielding approximately 300 µg of compound 29 (Purification workflow and purified HPLC-DAD peak, Supplementary Figs S81 and S82).
Confirmation of all the purified peaks was performed by using analytical column Ascentis® Express C18. Solvent B: ACN, solvent A: water, both solvents were HPLC grade. Conditions: 0–1 min 5% B, 1–16 min 5–100% B, 16–18 min 100% B, 18–20 min 100–5%, total run 21 min. Flow rate of 1 mL/min.
NMR data of purified compounds
Compound 19: NMR (Supplementary Table S1 and Supplementary Figs S37–S40) and MS (HR-ESI-MS m/z 487.1590 (calcd for C25H27O10, [M + H]+, 487.1599) data are consistent with the structure of aquayamycin72.
Compound 20: NMR (Supplementary Table S2 and Supplementary Figs S42–S45) and MS (HR-ESI-MS m/z 453.1548 (calcd for C25H25O8, [M + H]+, 453.1544) data are consistent with the structure of urdamycinone B76.
Compound 21: NMR (Supplementary Table S3 and Supplementary Figs S47–S50) and MS (HR-ESI-MS m/z 451.1381 (calcd for C25H23O8, [M + H]+, 451.1387) data are consistent with the structure of galtamycinone79.
Compound 22: NMR (Supplementary Table S4 and Supplementary Figs S53–S56) and MS (HR-ESI-MS m/z 435.1457 (calcd for C25H23O7, [M + H]+, 435.1438) data are consistent with the structure of dehydroxyaquayamycin80.
Compound 23 (m/z 549 [M + H]+) (see Supplementary Table S5 and Supplementary Figs S58–S62 for complete NMR data): 1H NMR (500 MHz, CDCl3) δ 12.65 (s, 1H, 8-OH), 11.42 (s, 1H, 1-OH), 8.33 (d, J = 8.5 Hz, 1H, 6-H), 8.15 (d, J = 8.5 Hz, 1H, 5-H), 7.92 (d, J = 8.0 Hz, 1H, 10-H), 7.90 (d, J = 8.0 Hz, 1H, 11-H), 7.27 (s, 1H, 4-H), 7.16 (s, 1H, 2-H), 4.94 (d, J = 10.0 Hz, 1H, 1′-H), 4.25 (q, J = 6.5 Hz, 1H, 5″-H), 3.84 (m, 1H, 3′-H), 3.69 (s, 1H, 4″-H), 3.57 (m, 1H, 5′-H), 3.09 (t, J = 8.6 Hz, 1H, 4′-H), 2.56 (dd, J = 13.0, 4.6 Hz, 1H, 2′-Ha), 2.10 (m, 1H, 2″-Hb), 2.04 (m, 1H, 3″-Hb), 1.82 (m, 1H, 3″-Ha), 1.66 (m, 1H, 2″-Ha), 1.46 (m, 1H, 2′-Hb), 1.39 (d, J = 6.0 Hz, 3 H, 6′-H3), 1.27 (d, J = 6.6 Hz, 3H, 6″-H3). HR-ESI-MS m/z 549.2115 (calcd for C31H33O9, [M + H]+, 549.2119). Our NMR data, combined with MS data support the annotation of marangucycline A2.
Compound 29 (See Supplementary Table S6 and Figs S83–S90 for complete NMR data): 1H NMR (500 MHz, MeOH-d4) δ 7.11 (d, J = 15.7 Hz, 1H), 6.19 (d, J = 15.7 Hz, 1H), 6.14 (s, 1H), 5.73 (d, J = 10.3 Hz, 1H), 5.57 (s, 1H), 3.87 (s, 1H), 3.58 (br quint, 6.6, 6.3 Hz), 2.57 (dquint, J = 10.3, 6.7 Hz, 1H), 1.87 (s, 1H), 1.29 (s, 1H), 1.13 (d, J = 6.3 Hz, 1H), 1.05 (d, J = 6.7 Hz, 1H). 13C assignments through gHSQC and gHMBC (1H 500 MHz, 13C 125 MHz, MeOH-d4) δ 173.7, 166.5, 160.7, 141.4, 134.0, 117.6, 101.4, 143.5, 88.5, 56.6, 72.1, 41.9, 12.1, 21.2, 16.5. HR-ESI-MS m/z 265.1440 (calcd for C15H21O4, [M + H]+, 265.1434). The proposed structure corresponds to a new fungal compound, (E)-2-((2Z,5E,7Z)-2-methoxy-7,9,10-trimethyl-9,10-dihydro-4H-oxecin-4-ylidene) acetic acid.
Sample preparation for LC-MS/MS
After the incubation time of microbial interactions -inter-kingdom interactions- on solid media (96 h), the region of interest was excised from the Petri dish for extraction procedures. The removed region containing the microbial colonies of interest (mono- or co-cultures) was transferred to Eppendorf tubes and 1 mL of extraction solvent was added. Four solvent mixtures were used, each one for each of the four replicates of the mono- and co-culture samples: (1:1 acetonitrile:methanol (ACN:MeOH); 1:1 ACN:water; 1:1 ACN:MeOH 0.1% formic acid, 1:1 ACN:water 0.1% formic acid). After 10 minutes of sonication, the supernatant was transferred to a clean vial, centrifuged at 16873 rcf (14000 rpm – radius of rotor ~76.865 mm Eppendorf Centrifuge 5418) for 15 minutes and 300 µL of the top extract were transferred to clean 0.5 mL 96-well polypropylene plates (Agilent Technologies Inc., Santa Clara, CA, USA) and sealed with Zone-Free Sealing Film (Excel Scientific) for LC-MS/MS analysis.
LC-MS/MS experiments
Crude extracts were analyzed by UPLC-HRMS-MS/MS on an Agilent 1290 UPLC using a Kinetex™ 50 mm × 2.1 mm C18 RP column (1.7 µm particle size) coupled to a MicrOTOF-QII mass spectrometer (Bruker Daltonics) equipped with the standard Apollo ESI source. Solvent B: ACN containing 0.1% formic acid, solvent A: water 0.1% formic acid, both solvents were of LC-MS grade. The gradient employed for chromatographic separation was 5% solvent B for 1 minute, linear gradient from 5% B to 95% B in 8 minutes, kept at 95% B for 2 minutes, back to 5% B in 1 minute and kept at 5% B for 1 minute to end up with total run time of 13 minutes at a flow rate of 0.5 mL/min. MS spectra were acquired in positive ion mode in the range of 50–2000 m/z. External calibration was performed prior to data collection using ESI-L Low Concentration Tuning Mix (Agilent Technologies). Hexakis (2,2-difluoroethoxy) phosphazene (Synquest Laboratories) m/z 622.028960, used for internal calibration, was added into a calibrant reservoir and placed inside the ion source. Other instruments settings were as follows: capillary voltage 4000 V, nebulizer gas pressure (N2) 2.0 bar, ion source temperature 200 °C, dry gas flow 9 L/min source temperature, spectral rate 3 Hz for MS1 and 10 Hz for MS2. For acquiring MS/MS fragmentation, 10 most intense ions per MS1 were selected for subsequent CID with stepped CID energy applied. More detailed parameters for tandem MS were used as previously published85.
Molecular networking
Input data for molecular networking was generated through conversion of the LC-MS/MS raw data to.mzXML data format by using the Bruker Daltonics Software. Data were submitted to the molecular networking workflow at the GNPS platform (gnps.ucsd.edu)27. The public dataset of this work is available at ftp://massive.ucsd.edu/MSV000079048. Molecular networking output was imported and visualized by using Cytoscape86, version 8.3. The complete analysis can be accessed via: http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=5dd7b0a5aeca4354945bf5bd1bddac27.
The GNPS molecular network was created using the parameters as follows: the data was filtered by removing all MS/MS peaks within +/− 17 Da of the precursor m/z, then clustered with MS-Cluster with a parent mass tolerance of 0.5 Da and a MS/MS fragment ion tolerance of 0.5 Da to create consensus spectra. Further, only consensus spectra that contained 2 nearly identical spectra were considered. A network was then created where edges were filtered to have a cosine score above 0.7 and 6 or more matched peaks. Further edges between two nodes were kept in the network if and only if each of the nodes appeared in each other’s respective top 10 most similar nodes. The spectra in the network were then searched against GNPS’s spectral libraries. The library spectra were filtered in the same manner as the input data. All matches kept between network spectra and library spectra were required to have a score above 0.7 and at least 6 matched peaks. A total of 15676 spectra were considered corresponding to 1590 nodes, and 175 clusters containing 931 clusternodes in the molecular network.
Minimal Inhibitory Concentration (MIC) assay against Coniochaeta sp . FLe4
MIC assay involving compound 23 and amphotericin B (1) was carried out according to a serial dilution87. The tested substance was prepared by serial dilutions to half of the concentration of the previous well, from a range of concentrations between 400 and 0.1953 µg/mL.
Diffusion assay with purified compounds against Coniochaeta sp. FLe4
A chemical complementation assay with purified compounds was carried out. Briefly, 1 μL of a pre-culture of Coniochaeta sp. FLe4 in ISP-2 liquid media, as described in the Strains and culture conditions section, was spotted on Petri dishes (10 mm) containing 10 mL of ISP-2 agar media. A second spot of the solubilized compound at the required concentration was pipetted onto the plate. Plates were incubated at 30 °C for six days.
References
Freeman, E. M. The Seed-Fungus of Lolium temulentum, L., the Darnel. Philos. T. Roy. Soc. B. 1–27, doi:10.1098/rspl.1902.0057 (1904).
Gunatilaka, A. A. L. Natural products from plant-associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 69, 509–526, doi:10.1021/np058128n (2006).
Wilson, D. Endophyte - The evolution of a term, and clarification of its use and definition. Oikos 73, 274–276, doi:10.2307/3545919 (1995).
Schulz, B. & Boyle, C. The endophytic continuum. Mycol. Res. 109, 661–686, doi:10.1017/s095375620500273x (2005).
Ludwig-Muller, J. Plants and endophytes: equal partners in secondary metabolite production? Biotechnol. Lett. 37, 1325–1334, doi:10.1007/s10529-015-1814-4 (2015).
Newman, D. J. & Cragg, G. M. Endophytic and epiphytic microbes as “sources” of bioactive agents. Front. Chem. 3, 1–13, doi:10.3389/fchem.2015.00034 (2015).
Stierle, A., Strobel, G. & Stierle, D. Taxol and taxane production by taxomyces andreanae, and endophytic fungus of Pacific Yew. Science 260, 214–216, doi:10.1126/science.8097061 (1993).
Kusari, S., Lamshoeft, M. & Spiteller, M. Aspergillus fumigatus Fresenius, an endophytic fungus from Juniperus communis L. Horstmann as a novel source of the anticancer pro-drug deoxypodophyllotoxin. J. Appl. Microbiol. 107, 1019–1030, doi:10.1111/j.1365-2672.2009.04285.x (2009).
Kusari, S. et al. Endophytes are hidden producers of maytansine in Putterlickia Roots. J. Nat. Prod. 77, 2577–2584, doi:10.1021/np500219a (2014).
Kusari, P. et al. Cross-species biosynthesis of maytansine in Maytenus serrata. RSC Advances 6, 10011–10016, doi:10.1039/c5ra25042k (2016).
Venugopalan, A., Potunuru, U. R., Dixit, M. & Srivastava, S. Effect of fermentation parameters, elicitors and precursors on camptothecin production from the endophyte Fusarium solani. Bioresource Technol. 206, 104–111, doi:10.1016/j.biortech.2016.01.079 (2016).
Kusari, S., Zuehlke, S. & Spiteller, M. An Endophytic fungus from Camptotheca acuminata that produces camptothecin and analogues. J. Nat. Prod. 72, 2–7, doi:10.1021/np800455b (2009).
Gouda, S., Das, G., Sen, S. K., Shin, H. S. & Patra, J. K. Endophytes: A treasure house of bioactive compounds of medicinal importance. Front.Microbiol. 7, 8, doi:10.3389/fmicb.2016.01538 (2016).
Scherlach, K. & Hertweck, C. Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol. Chem. 7, 1753–1760, doi:10.1039/b821578b (2009).
Bertrand, S. et al. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 32, 1180–1204, doi:10.1016/j.biotechadv.2014.03.001 (2014).
Marmann, A., Aly, A. H., Lin, W. H., Wang, B. G. & Proksch, P. Co-Cultivation-A powerful emerging tool for enhancing the chemical diversity of microorganisms. Mar. Drugs. 12, 1043–1065, doi:10.3390/md12021043 (2014).
Rutledge, P. J. & Challis, G. L. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523, doi:10.1038/nrmicro3496 (2015).
Bertrand, S. et al. Detection of metabolite induction in fungal co-cultures on solid media by high-throughput differential ultra-high pressure liquid chromatography-time-of-flight mass spectrometry fingerprinting. J. Chromatogr. A 1292, 219–228, doi:10.1016/j.chroma.2013.01.098 (2013).
Traxler, M. F., Watrous, J. D., Alexandrov, T., Dorrestein, P. C. & Kolter, R. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. Mbio 4, 12, doi:10.1128/mBio.00459-13 (2013).
Chagas, F. O., Dias, L. G. & Pupo, M. T. A mixed culture of endophytic fungi increases production of antifungal polyketides. J. Chem. Ecol. 39, 1335–1342, doi:10.1007/s10886-013-0351-7 (2013).
Zhu, F., Chen, G. Y., Wu, J. S. & Pan, J. H. Structure revision and cytotoxic activity of marinamide and its methyl ester, novel alkaloids produced by co-cultures of two marine-derived mangrove endophytic fungi. Nat. Prod. Res. 27, 1960–1964, doi:10.1080/14786419.2013.800980 (2013).
Soliman, S. S. M. & Raizada, M. N. Interactions between co-habitating fungi elicit synthesis of Taxol from an endophytic fungus in host Taxus plants. Front. Microbiol. 4, 14, doi:10.3389/fmicb.2013.00003 (2013).
Kelsic, E. D., Zhao, J., Vetsigian, K. & Kishony, R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature 521, 516–U208, doi:10.1038/nature14485 (2015).
Talbot, N. J. Plant Immunity: A little help from fungal friends. Curr. Biol. 25, R1074–R1076, doi:10.1016/j.cub.2015.09.068 (2015).
Soliman, S. S. M. et al. An Endophyte constructs fungicide-containing extracellular barriers for its host plant. Curr. Biol. 25, 2570–2576, doi:10.1016/j.cub.2015.08.027 (2015).
Watrous, J. et al. Mass spectral molecular networking of living microbial colonies. P. Natl. Acad. Sci. USA 109, E1743–E1752, doi:10.1073/pnas.1203689109 (2012).
Wang, M. X. et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34, 828–837, doi:10.1038/nbt.3597 (2016).
Yang, J. Y. et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 76, 1686–1699, doi:10.1021/np400413s (2013).
Allard, P. M. et al. Integration of molecular networking and In-Silico MS/MS fragmentation for natural products dereplication. Anal. Chem. 88, 3317–3323, doi:10.1021/acs.analchem.5b04804 (2016).
Nguyen, D. D. et al. Indexing the Pseudomonas specialized metabolome enabled the discovery of poaeamide B and the bananamides. Nat. Microbiol. 2, 9, doi:10.1038/nmicrobiol.2016.197 (2017).
Allard, P. M., Genta-Jouve, G. & Wolfender, J. L. Deep metabolome annotation in natural products research: towards a virtuous cycle in metabolite identification. Curr. Opin. Chem. Biol. 36, 40–49, doi:10.1016/j.cbpa.2016.12.022 (2017).
Borella, J. C., Lopes, J. L. C., Vichnewski, W., Cunha, W. R. & Herz, W. Sesquiterpene lactones, triterpenes and flavones from Lychnophora ericoides and Lychnophora pseudovillosissima. Biochem. Syst. Ecol. 26, 671–676, doi:10.1016/s0305-1978(98)00004-0 (1998).
Borsato, M. L. C., Grael, C. F. F., Souza, G. E. P. & Lopes, N. P. Analgesic activity of the lignans from Lychnophora ericoides. Phytochemistry 55, 809–813, doi:10.1016/s0031-9422(00)00388-5 (2000).
Sakamoto, H. T. et al. Sesquiterpene lactones from Lychnophora ericoides. J. Nat. Prod. 66, 693–695, doi:10.1021/np020314v (2003).
dos Santos, P. A. et al. Production of an antiproliferative furanoheliangolide by Lychnophora ericoides cell culture. Chem. Pharm. Bull. 52, 1433–1435 (2004).
dos Santos, M. D., Gobbo-Neto, L., Albarella, L., de Souza, G. E. P. & Lopes, N. P. Analgesic activity of di-caffeoylquinic acids from roots of Lychnophora ericoides (Arnica da serra). J. Ethnopharmacol. 96, 545–549, doi:10.1016/j.jep.2004.09.043 (2005).
Gobbo-Neto, L. et al. Evaluation of the anti-inflammatory and antioxidant activities of di-C-glucosylflavones from Lychnophora ericoides (Asteraceae). Planta Med. 71, 3–6, doi:10.1055/s-2005-837742 (2005).
Gobbo-Neto, L. et al. Glycosides, caffeoylquinic acids and flavonoids from the polar extract of leaves of Lychnophora ericoides Mart. (Asteraceae). Biochem. Syst. Ecol. 36, 473–475, doi:10.1016/j.bse.2007.12.002 (2008).
Gobbo-Neto, L., Gates, P. J. & Lopes, N. P. Negative ion ‘chip-based’ nanospray tandem mass spectrometry for the analysis of flavonoids in glandular trichomes of Lychnophora ericoides Mart. (Asteraceae). Rapid Commun. Mass Sp 22, 3802–3808, doi:10.1002/rcm.3802 (2008).
Gobbo-Neto, L. & Lopes, N. P. Online identification of chlorogenic acids, sesquiterpene lactones, and flavonoids in the Brazilian arnica Lychnophora ericoides Mart. (Asteraceae) leaves by HPLC-DAD-MS and HPLC-DAD-MS/MS and a validated HPLC-DAD method for their simultaneous analysis. J. Agr. Food Chem. 56, 1193–1204, doi:10.1021/jf0728121 (2008).
Gobbo-Neto, L. et al. Differential metabolic and biological profiles of Lychnophora ericoides Mart. (Asteraceae) from different localities in the Brazilian “campos rupestres”. J. Brazil. Chem. Soc. 21, 750–759, doi:10.1590/s0103-50532010000400023 (2010).
Keles, L. C. et al. Lychnophorinae (Asteraceae): A survey of its chemical constituents and biological activities. Quim. Nova 33, 2245–2260, doi:10.1590/s0100-40422010001000038 (2010).
Semir, J., Rezende, A. R., Monge, M. & Lopes, N. P. As Arnicas Endêmicas das Serras do Brasil [The endemic Arnicas of Brazilian Hill tops]. First Edt. edn (2011).
Conti, R. et al. Endophytic actinobacteria from the Brazilian medicinal plant Lychnophora ericoides Mart. and the biological potential of their secondary metabolites. Chem. Biodivers. 13, 727–736, doi:10.1002/cbdv.201500225 (2016).
Chagas, F. O., Caraballo-Rodríguez, A. M., Dorrestein, P. C. & Pupo, M. T. Expanding the chemical repertoire of the endophyte Streptomyces albospinus RLe7 reveals amphotericin B as inducer of a fungal phenotype. J. Nat. Prod. 80, 1302–1309, doi:10.1021/acs.jnatprod.6b00870 (2017).
Nguyen, D. D. et al. MS/MS networking guided analysis of molecule and gene cluster families. P. Natl. Acad. Sci.USA 110, E2611–E2620, doi:10.1073/pnas.1303471110 (2013).
Chen, Y., Peng, Z. H., Song, W., Zhu, M. M. & Han, F. M. Chromatographic tandem mass spectrometric detection of physostigmine and its major metabolites in rat urine. Anal. Lett. 40, 3256–3266, doi:10.1080/00032710701672749 (2007).
Shirafuji, H., Tsuboya, S. & Harada, S. Acetylcholinesterase inhibitors TAN-1169 and their manufacture with Streptomyces species. Japan patent JP 02270875 A (1990).
Liu, J., Ng, T., Rui, Z., Ad, O. & Zhang, W. J. Unusual acetylation-dependent reaction cascade in the biosynthesis of the pyrroloindole drug physostigmine. Angew. Chem. Int. Edit 53, 136–139, doi:10.1002/anie.201308069 (2014).
Murao, S. & Hayashi, H. Physostigmine and N-8-norphysostigmine, insecticidal compounds, from Streptomyces sp. Agr. Biol. Chem. Tokyo 50, 523–524 (1986).
Ulrych, A. et al. Dissociation of nystatin and amphotericin analogues: characterisation of minor anti-fungal macrolides. Eur. J. Mass Spectrom. 16, 73–80, doi:10.1255/ejms.1027 (2010).
Sowinski, P., Kolodziejczyk, P. & Borowski, E. The structure of amphotericin A. 1. Mass spectrometric analysis of dodecahydroamphotericin A. J. Antibiot. 38, 169–174 (1985).
Stiller, E. T., Vandeputte, J. & Wachtel, J. L. Amphotericins A and B, antifungal antibiotics produced by a streptomycete. II. The isolation and properties of the crystalline amphotericins. Antibiotics annual 3, 587–591 (1955).
Li, J., Zhu, H. Q., Li, J. Y., Jin, S. H. & Hu, C. Q. Isolation, structure elucidation and activity of an unknown impurity of amphotericin B. J. Antibiot. 60, 272–276 (2007).
Palaniswamy, V. A. et al. Preparation and structural elucidation of amphotericin X. Planta Med. 56, doi:10.1055/s-2006-961145 (1990).
Byrne, B., Carmody, M., Gibson, E., Rawlings, B. & Caffrey, P. Biosynthesis of deoxyamphotericins and deoxyamphoteronolides by engineered strains of Streptomyces nodosus. Chem. Biol. 10, 1215–1224, doi:10.1016/j.chembiol.2003.12.001 (2003).
Anderson, T. M. et al. Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol. 10, 400–U121, doi:10.1038/nchembio.1496 (2014).
Kim, I. S. & Lee, K. J. Regulation of production of leupeptin, leupeptin-inactivating enzime, and trypsin-like protease in Streptomyces-exfoliatus SMF13. J. Ferment. Bioeng. 80, 434–439, doi:10.1016/0922-338x(96)80916-0 (1995).
Aoyagi, T. et al. Leupeptins, new protease inhibitors from actinomycetes. J. Antibiot. 22, 283–286 (1969).
Kondo, S. et al. Isolation and characterization of leupeptins produced by actinomycetes. Chem. Pharm. Bull. 17, 1896–1901 (1969).
Hamato, N., Takano, R., Kameihayashi, K., Imada, C. & Hara, S. Leupeptins produced by the marine Alteromonas sp. B-10-31. Biosci. Biotech. Bioch. 56, 1316–1318 (1992).
McConnell, R. M., York, J. L., Frizzell, D. & Ezell, C. Inhibition studies of some serine and thiol proteinases by new leupeptin analogues. J. Med. Chem. 36, 1084–1089, doi:10.1021/jm00060a016 (1993).
Liu, Q.-y, Liu, H.-z, Chen, W. & Chi, Z.-w The component separation and structure of thrombin inhibitor produced by Streptomyces S-254. Zhongguo Kangshengsu Zazhi 15, 342–346, doi:10.13461/j.cnki.cja.001361 (1990).
Liu, H.-z, Chen, W. & Liu, Q.-y Fermentation, extract and the activity of thrombin inhibitor in Streptomyces S-254. Zhongguo Kangshengsu Zazhi 15, 407–411, doi:10.13461/j.cnki.cja.001376 (1990).
Ogura, K. et al. Purification and structure of a novel cysteine proteinase-inhibitor, strepin P-1. Agr. Biol. Chem. Tokyo 49, 799–805 (1985).
Kim, I. S. & Lee, K. J. Physiological roles of leupeptin and extracellular proteases in mycelium development of Streptomyces-exfoliatus smf13. Microbiology-Uk 141, 1017–1025 (1995).
Kim, D. W. et al. Proteases and protease inhibitors produced in streptomycetes and their roles in morphological differentiation. J. Microbiol. Biotechn. 16, 5–14 (2006).
Song, Y. X. et al. Cytotoxic and antibacterial angucycline- and prodigiosin- analogues from the deep-sea derived Streptomyces sp. SCSIO 11594. Mar. Drugs 13, 1304–1316, doi:10.3390/md13031304 (2015).
Oki, T. et al. Antitumor anthracycline antibiotics, aclacinomycin a and analogues. II. Structural determination. J. Antibiot. 32, 801–819 (1979).
Ayukawa, S. et al. Inhibition of tyrosine hydroxylase by Aquayamycin. J. Antibiot. 21, 350–353 (1968).
Sezaki, M. et al. Studies on an new antibiotic pigment Aquayamycin. J. Antibiot. 21, 91–97 (1968).
Sezaki, M., Kondo, S., Maeda, K., Umezawa, H. & Ohno, M. Structure of Aquayamycin. Tetrahedron 26, 5171–5190, doi:10.1016/s0040-4020(01)98726-5 (1970).
Kazutoshi, S. New Antibiotic Substance DQ112-A, Production thereof and Antitumor Agent. Japan patent 07–324082 (1995).
Takayoshi, O. et al. Antitumor Substance BA-12100S, Production and its use. Japan patent 01–278183 (1990).
Dictionary of Natural Products http://dnp.chemnetbase.com/ (2017)
Rohr, J. & Zeeck, A. Metabolic products of microorganisms. 240. Urdamycins, new angucycline antibiotics from Streptomyces fradiae. 2. Structural studies of urdamycin B to urdamycin F. J. Antibiot. 40, 459–467 (1987).
Apsel, B. et al. General entries to C-aryl glycosides. Formal synthesis of galtamycinone. Tetrahedron Lett. 44, 1075–1077, doi:10.1016/s0040-4039(02)02666-7 (2003).
Murenets, N. V., Kudinova, M. K., Klyev, N. A., Chernyshev, A. I. & Shorshnev, S. V. Galtamycin - Galtamycinone Structure. Antibiotiki I Meditsinskaya Biotekhnologiya 31, 431–434 (1986).
Stroch, K., Zeeck, A., Antal, N. & Fiedler, H. P. Retymicin, galtamycin B, saquayamycin Z and ribofuranosyllumichrome, novel secondary metabolites from Micromonospora sp Tu 6368 - II. Structure elucidation. J. Antibiot. 58, 103–110 (2005).
Supong, K. et al. Antimalarial and antitubercular C-glycosylated benz alpha anthraquinones from the marine-derived Streptomyces sp. BCC45596. Phytochem. Lett. 5, 651–656, doi:10.1016/j.phytol.2012.06.015 (2012).
Kharel, M. K. et al. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 29, 264–325, doi:10.1039/c1np00068c (2012).
Wang, W. S. et al. Angucyclines as signals modulate the behaviors of Streptomyces coelicolor. P. Natl. Acad. Sci. USA 111, 5688–5693, doi:10.1073/pnas.1324253111 (2014).
Okada, B. K., Wu, Y. H., Mao, D. N., Bushin, L. B. & Seyedsayamdost, M. R. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem. Biol. 11, 2124–2130, doi:10.1021/acschembio.6b00447 (2016).
Covington, B. C., McLean, J. A. & Bachmann, B. O. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat. Prod. Rep. 34, 6–24, doi:10.1039/c6np00048g (2017).
Garg, N. et al. Mass spectral similarity for untargeted metabolomics data analysis of complex mixtures. Int. J. Mass Spectrom. 377, 719–727, doi:10.1016/j.ijms.2014.06.005 (2015).
Shannon, P. et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504, doi:10.1101/gr.1239303 (2003).
CLSI. In Approved Standard. Second Edition. CLSI Document MA38-A2 (Clinical and Laboratory Standards Institute, 950 West Valley Road, Suite 2500, Wayne, Pennsylvania, 19087, USA, 2008).
Acknowledgements
AMCR would like to thank Fernanda O. das Chagas for improving the manuscript and NMR discussions, Thomas Hoffman, Vanessa Phelan and Gleb Pishchany for their comments and suggestions on the manuscript, and Dorrestein lab´ members for their support in MS approaches. This work was supported by grants#2012/21803-1, 2014/01651-8 and 2008/09540-0 São Paulo Research Foundation (FAPESP), CEPID-CIBFar (FAPESP 2013/07600-3) and the National Institute of Health (NIH) (grant numbers 5P41GM103484, GMS10RR029121). Authors also thank University of California – San Diego (UCSD), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and INCT-INBEQMeDI for supporting research.
Author information
Authors and Affiliations
Contributions
A.M.C.R., P.C.D. and M.T.P. designed the research, A.M.C.R. performed experiments, A.M.C.R., P.C.D. and M.T.P. analyzed data, discussed results and contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Caraballo-Rodríguez, A.M., Dorrestein, P.C. & Pupo, M.T. Molecular inter-kingdom interactions of endophytes isolated from Lychnophora ericoides . Sci Rep 7, 5373 (2017). https://doi.org/10.1038/s41598-017-05532-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-05532-5
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.