Abstract
The fundamental trait in selective breeding of oil palm (Eleais guineensis Jacq.) is the shell thickness surrounding the kernel. The monogenic shell thickness is inversely correlated to mesocarp thickness, where the crude palm oil accumulates. Commercial thin-shelled tenera derived from thick-shelled dura × shell-less pisifera generally contain 30% higher oil per bunch. Two mutations, sh MPOB (M1) and sh AVROS (M2) in the SHELL gene – a type II MADS-box transcription factor mainly present in AVROS and Nigerian origins, were reported to be responsible for different fruit forms. In this study, we have tested 1,339 samples maintained in Sime Darby Plantation using both mutations. Five genotype-phenotype discrepancies and eight controls were then re-tested with all five reported mutations (sh AVROS, sh MPOB, sh MPOB2, sh MPOB3 and sh MPOB4) within the same gene. The integration of genotypic data, pedigree records and shell formation model further explained the haploinsufficiency effect on the SHELL gene with different number of functional copies. Some rare mutations were also identified, suggesting a need to further confirm the existence of cis-compound mutations in the gene. With this, the prediction accuracy of fruit forms can be further improved, especially in introgressive hybrids of oil palm. Understanding causative variant segregation is extremely important, even for monogenic traits such as shell thickness in oil palm.
Similar content being viewed by others
Introduction
Elaeis, the only genus under the Arecaceae family, produces edible oil. Elaeis guineensis Jacq. of West African origin is the predominantly used species, planted due to oil yield superiority, whereas the planting of another related species, E. oleifera (HBK) Cortes is usually restricted to Latin America, where the species originated from. In oil palm, three naturally occurring fruit forms (dura, tenera and pisifera) can be recognized based on their shell thickness, which is controlled by a single gene with two co-dominant alleles1. The thinner shell usually confers additional mesocarp to the fruit, allowing more oil to accumulate. Hence, planters in Southeast Asia in the 1960s switched from planting Deli dura (thick-shelled) fruit to tenera (thin-shelled) fruit by making hybrids between Deli dura and AVROS pisifera (shell-less, typically female sterile), realizing a 30% increment in oil/hectare2, 3.
Two independent mutations in the SHELL gene – a type II MADS-box transcription factor which is homologous with Arabidopsis SEEDSTICK (STK) and rice OsMADS13 – were recently reported to be responsible for the fruit forms4. The two mutations, sh MPOB (T → C, for leucine → proline amino acid change) and sh AVROS (A → T, for lysine → asparagine amino acid change) were identified among the descendants of a Nigerian tenera accession and a Congo-derived AVROS pisifera accession, respectively4. These mutations result in amino acid changes within the MADS-box domain and disrupt heterodimerization (sh MPOB) and DNA binding (sh AVROS)4, 5, respectively. The homozygous wild-type sh DeliDura nucleotides at both variant positions are carried by dura palms. Palms which are heterozygous for either of the two mutations (sh DeliDura/sh MPOB or sh DeliDura/sh AVROS) produce tenera palms. Pisifera palms are homozygous for either mutation or heteroallelic for both mutations. In other words, the most obvious causative mechanism for the co-dominant fruit form trait is mainly due to haploinsufficiency, wherein only a single functional copy of SHELL gene in a diploid genome is insufficient to maintain the wild-type function i.e. dura.
From the mass screening of commercial tenera materials, another three non-synonymous variants (sh MPOB2, sh MPOB3 and sh MPOB4) within the same MADS-box coding regions controlling the fruit form phenotype in a similar manner were then identified6. An example of potential use would be for pisifera breeding, where both pisifera and tenera are assessed, but dura discarded. The markers developed could be used to remove the undesired dura seeds derived from tenera × tenera crosses at the pre-nursery stage. This will improve both genetic gain over generations and trial management through reduced land requirement. Also, the SHELL genetic testing could potentially increase the oil yield of planted areas by eliminating the lower-yielding, non-tenera contaminants prior to field planting6. The reported mutations in the SHELL gene are applicable in Nigerian and the commercial AVROS accessions, separately. However, some breeding institutes are aiming to broaden the genetic base of AVROS accessions through introgression, or have switched to other genetic resources for paternal lineages, such as Ekona, Yangambi, LaMe and Serdang fertile pisifera (SP). A better understanding of the heterozygous action of the loss-of-function mutations in SHELL gene is also essential to further improve fruit form predictability, especially for introgressive hybrids of oil palm.
Result and Discussion
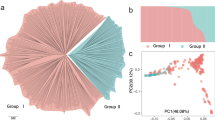
In this study, we carried out SHELL genetic testing on Sime Darby Plantation’s repertoire of genetic materials using the sh MPOB and sh AVROS tests. The objectives were to validate the SHELL markers on these materials and understand the effect of introgression from different pisifera sources on fruit form prediction. As the first phase of validation, three fruit forms (120 dura, 305 tenera and 162 pisifera) belonging to four paternal stocks were selected (Table 1). The genetic stratification of the 587 palms was determined through principal component analysis (PCA) based on our OP200K genotyping array7. Four main clusters i.e. I) Ekona, II) AVROS, III) AVROS x SP, IV) AVROS-SP × Ekona (Fig. 1A) were identified and these showed a good agreement with the known pedigree information (Fig. 1B). The AVROS pisifera is selected for superiority in growth uniformity, precocity and mesocarp oil content when combined with Deli dura origins to produce commercial tenera planting materials. However, the narrow genetic base of AVROS (derived from only one palm selected from the self-pollination of the SP540 palm which was again self-pollinated, although there is evidence for some illegitimacy from the final self-pollination) may hinder future breeding progress in oil palm8. Oil palm breeders, hence, are actively looking for other genetic resources to enrich the genetic diversity of the current AVROS accession (cluster II). One of these is SP, which was initially developed in the 1960’s for shell-less fertile pisifera planting materials9, but seed germination and oil extraction from pisifera fruits still remains challenging2. By having AVROS × SP (cluster III), the breeders aimed to further reduce shell thickness in the commercial tenera without complete loss of the shell. Another examined option was the Ekona origin (cluster I) which originated from Cameroon. The trial result showed that Ekona pisifera gave smaller trunk height increments, as good or even better bunch production and oil/bunch than the AVROS pisifera 10, 11. In Sime Darby, we have further introgressed cluster I into cluster III to generate AVROS-SP × Ekona hybrids (cluster IV) (Fig. 1B). This relationship was clearly reflected as cluster IV was located in between cluster I and cluster III (Fig. 1A).
Genetic stratification of 587 oil palm samples, as the first validation set representing four paternal stocks. (A) Four clusters i.e. I) Ekona, II) AVROS, III) AVROS x SP, and IV) AVROS-SP × Ekona corresponding to their known origins were detected. PC1 and PC2 indicate the scores of principle components 1 and 2, respectively. (B) A pedigree of the four origins. SP – Serdang fertile pisifera.
The fruit forms of individual palms were phenotyped based on visual inspection of shell thickness and the diagnostic presence of a fiber ring around the kernel in the tenera fruit. To validate the accuracy of SHELL markers, the fruit form data was compared to the genotype data. The two mutations were defined as M1 for sh MPOB and M2 for sh AVROS, respectively. In general, the prediction accuracy of the SHELL markers was successfully validated (100% accuracy), except in the AVROS-SP × Ekona accession (Fig. 2) (Supplementary Table 1). The AVROS accession and the Ekona accession carried sh AVROS and sh MPOB mutations, respectively, which may have arisen independently in these accessions derived from central and western Africa. The introgressive hybridization between AVROS × SP and Ekona origins introduced polymorphism for both M1 and M2 (Fig. 2) (Supplementary Table 1) into their progenies. Using single marker prediction, neither M1 nor M2 correctly predicted the segregating dura, tenera and pisifera fruit forms. We then expanded the SHELL testing using a second validation set with a total of 752 palms representing AVROS, MPOB, Congo, Tanzanian and four introgressive hybrid origins (AVROS × Nigerian, Cameroon × Congo, Deli-Nigerian × URT-Calabar and URT × Calabar) (Table 2) (Supplementary Table 2). The same result was observed, where the M2 was carried by Congo and Tanzanian originated from central and eastern Africa. The fruit forms of the pure accessions (except AVROS accession with 72.00% of accuracy) thus can be predicted accurately using a single marker, but not in the introgressed hybrids (Table 2) (Supplementary Table 2).
To interpret this further, we mapped all the genotype-phenotype relationships onto an AVROS-SP tenera × Ekona tenera hybrid population (Fig. 3). The AVROS-SP tenera containing a single heterozygous mutation at M2 (sh DeliDura/sh DeliDura; sh DeliDura/sh AVROS) was hybridized to the Ekona tenera that carried the other single heterozygous mutation at M1 (sh DeliDura/sh MPOB; sh DeliDura/sh DeliDura), and the cross produced four genotypes in the hybrid population, according to Mendelian inheritance. In this population, we confirmed the presence of the haploinsufficiency effect on the SHELL gene. Not all loss-of-function mutations of an encoded protein are deleterious. Some heterozygous loss-of-function mutations, mainly due to haploinsufficiency, even give selective advantages12. The scenario was clearly demonstrated in thin-shelled tenera, which produce 30% more oil with full fertility compared to their dura and pisifera parents. The tenera palms in this study contained only a single functional copy of a gene per diploid genome causing an inability to produce enough functional protein as wild-type dura palms in the cells; this leads to a haploinsufficiency of the heterodimer which mediates the effects of the SHELL gene. Hence, we scored the wild-type sh DeliDura/sh DeliDura which occurred at both M1 and M2 as two functional copies; one functional copy for a single mutation at either the M1 or M2 variant position per diploid; and nil functional copy for multiple mutations at both M1 and M2 positions. Next, we link the haploinsufficiency effect to the shell formation model through SHELL-SEP-like protein heterodimerization and DNA binding4,5,6 (Fig. 3). The dura palms with two functional copies were not disrupted in shell formation at all, as expected. As for tenera, the two genotypes possible only contributed one functional copy of the SHELL protein, which explained the thinner shell compared to dura. One of the alleles led to an inactive protein heterodimer due to failure in either DNA binding for sh AVROS carriers, or SHELL-SEP-like dimerization itself for sh MPOB carriers. All the shell-less pisifera palms were heteroallelic for both mutations (nil functional copy). Therefore, it is important to determine the polymorphism at M1 and M2 first to decide which prediction method should be used. Here, we can observe the clear effect of the causal variants influencing the shell thickness when introgression between certain populations takes place. By shifting single marker analysis to functional copy counting, the prediction accuracies of fruit form in the four introgressive hybrids were significantly improved, ranging from 96.00 to 100.00% (Table 2). Nevertheless, we could not observe any association between both mutations and fertile pisifera (with the availability of the four fertile pisifera) in AVROS-SP accessions, indicating that the SHELL gene itself might not be responsible solely for female sterility in pisifera (Supplementary Table 1).
The mapping of genotype-phenotype relationships on an AVROS-SP tenera × Ekona tenera hybrid population and SHELL-SEP-like protein interaction model in three fruit forms influenced by functional copy (haploinsufficiency effect). The pisifera fruit form was due to trans-compound heterozygosity in SHELL gene.
The global accuracy in the second validation set was extremely high (99.34%), but unexpected low accuracy (72.00%) in one AVROS accession drew our attention (Table 2). The phenotype-genotype discrepancies might be due to the presence of other novel mutants on the SHELL gene, as reported6. Hence, a total of 13 palms (5 phenotype-genotype discrepancies and 8 controls) were tested using all five reported mutant SHELL alleles including sh AVROS, sh MPOB, sh MPOB2, sh MPOB3 and sh MPOB4 within the MADS-box domain. The three AVROS discrepancies were successfully rectified based on sh MPOB2 and sh AVROS mutations (Table 3), achieving 100.00% prediction accuracy in the accession. However, this did not apply to two Cameroon × Congo discrepancies, indicating probable occurrence of other novel mutations (Table 3). We thus expect another heterozygous mutation for V599 pisifera and V927 tenera. Interestingly, we also discovered a heteroallelic case for three mutations in V121 pisifera in Nigerian × AVROS accession. This implied that some accessions/origins can possess more than two mutations or a different cluster of mutations, although it can be rare. However, this will become prominent during introgression of wild germplasm. The compound mutation in the SHELL gene for fruit form phenotype in oil palm was firstly discussed in this study. Multiple mutations that occur in the same gene and in different genes, are defined as compound mutation and double mutation, respectively13, 14 (Fig. 4). All the reported mutations are located in the SHELL gene, therefore known as compound mutations which can be heterozygous or homozygous. The loss-of-function compound mutations can happen in cis (mutations on the same chromosome) (Fig. 4B) and trans forms (mutations on the different chromosome) (Fig. 4C) too. To date, the haploinsufficient SHELL gene for pisifera is mainly explained by trans-compound heterozygous mutations only, particularly in introgressive hybrids. Nevertheless, we should not ignore the possible existence of cis-compound heterozygous palms that are still able to produce one functional copy to express the tenera fruit form (Fig. 4B). Hence, these individual palms can be misinterpreted as pisifera if based on heteroallelic tests for both mutations only.
The zygosity of loss-of-function mutations in SHELL gene. (A) Tenera palm that carried a single heterozygous mutation, only produce 1 functional copy of the gene. (B) Cis-compound heterozygous tenera palms produce 1 functional copy, when both mutations occur on the same chromosome. (C) Trans-compound heterozygous pisifera palms produce 0 functional copy, when both mutations occur on different chromosome. (D) In double heterozygous individuals, the mutations occurred in different genes.
In conclusion, the haploinsufficiency effect on SHELL gene responsible for fruit form variation in oil palm was further characterized based on identity-by-descent. The reported sh AVROS, sh MPOB, sh MPOB2, sh MPOB3 and sh MPOB4 within MADS-box domain should be adopted together to improve prediction accuracy, especially in the introgressive hybrid materials. Furthermore, the existence of cis-compound heterozygous palms should be confirmed through continuous screening of various germplasm. This study provides a good reference for other crop and animal breeding programs where the understanding of causative variant segregation is extremely important, even for monogenic traits such as oil palm shell thickness.
Methods
Sampling and DNA Preparation
A total of 1,339 oil palm samples were selected and divided into two validation sets. The first validation set included 587 palms representing four different accessions, i.e. AVROS, AVROS × SP, AVROS-SP × Ekona and Ekona maintained at Sime Darby Plantation R&D Centre in Malaysia (Table 1). The validation set consisted of three different fruit forms (thick-shelled dura, thin-shelled tenera and shell-less pisifera). The pedigree was provided by the breeders as indicated in Fig. 1B. Subsequently, the study was extended to the second validation set derived from more diverse backgrounds, also with a larger overall group size (752 palms) (Table 2). The background included four pure accessions (AVROS, MPOB, Tanzanian and Congo) and four introgressed hybrid populations (Nigerian × AVROS, Cameroon × Congo, Deli-Nigerian × URT Calabar and URT × Calabar). Total genomic DNA was isolated from 0.1 g of young leaf tissue (frond 0) using the DNAeasy Plant Mini Kit (Qiagen).
Genetic stratification analysis
To understand the genetic structure, the first validation set was genotyped using genome-wide SNP markers on a OP200K Infinium array7 (Illumina). The genotypic data of 200,000 SNPs were filtered based on minor allele frequency (MAF) >0.01 and >90% call rate. Subsequently, the genetic stratification among 586 oil palm samples was analyzed in the R package SNPRelate15, using the default parameters.
SHELL genotyping
About 3 ng of genomic DNA was used as template for both specific allele tests. The probe design for sh MPOB, sh AVROS, sh MPOB2, sh MPOB3 and sh MPOB4 mutations was done at their reported genomic positions: 3,078,161 bp, 3,078,154 bp, 3,078,180 bp, 3,078,125 bp and 3,078,178 bp on Chromosome 2, namely CM002082.14, 6 based on the requirement of Kompetitive Allele Specific PCRTM (KASPTM) genotyping platform. Two fluorophores FAM and HEX were used to distinguish the KASPTM genotyping data. In the clustering plot, the samples marked red were homozygous for alleles reported with HEX, whereas those marked blue were homozygous for FAM allele. The heterozygous samples appear as green.
Genotype-phenotype analysis
The fruit form census was performed based on visual examination of shell thickness and the presence or absence of a fiber ring in each cross-sectioned fruitlet. In addition, fertility status of AVROS-SP pisifera was also recorded based on the presence of embryo in the palm kernel. The observed fruit form and fertility data were then compared to the predicted data based on the reported sh MPOB and sh AVROS variants4. From there, accuracy (in %) of the single marker and haploinsufficiency models in each accession, except 15 samples with missing data were evaluated (Supplementary Table 1). The single marker method predicted the fruit forms based on either sh MPOB or sh AVROS variant. But, the haploinsufficiency model considered both variants simultaneously. The wild-type sh DeliDura occurred at both M1 and M2 per chromosome, with 2 functional copies of the M1-M2 haplotype for dura; a single mutation at either M1 or M2 per chromosome as 1 functional copy for tenera; and compound mutations at M1 and M2 on both haplotypes as 0 functional copy for pisifera. The analysis was then extended to another three mutations, including sh MPOB2, sh MPOB3 and sh MPOB4 tested on five discrepancies and eight controls based on the same haploinsufficiency model.
References
Beirnaert, A. & Vanderweyen, R. Contribution à l’étude génétique et biométrique des variétiés d’Elaeis guineensis Jacq. In Publ. Inst. Nat. Etude Agron. Congo Belge. Ser. Sci. Vol. 27 1–101 (1941).
Corley, R. H. V. & Tinker, P. B. Selection and breeding. In The Oil Palm 133–187 (Blackwell 2003).
Hardon, J. J., Corley, R. H. V. & Lee, C. H. Breeding and selecting the oil palm 63–81 (Academic Press, London 1987).
Singh, R. et al. The oil palm SHELL gene controls oil yield and encodes a homologue of Seedstick. Nature 500, 340–344 (2013).
Ordway, J. et al. Expression of sep-like genes for identifying and controlling palm plant shell phenotypes. Vol. WO2015010131A3 (Malaysia 2015).
Ooi, L. C. -L. et al. Non-tenera Contamination and the Economic Impact of SHELL Genetic Testing in the Malaysian Independent Oil Palm Industry. Frontiers in Plant Science 7 (2016).
Kwong, Q. B. et al. Development and Validation of a High-Density SNP Genotyping Array for African Oil Palm. Molecular Plant 9, 1132–1141 (2016).
Rosenquist, E. A. An overview of breeding technology and selection in Elaeis guineensis. In International Oil Palm Development Conference - Agriculture 5–25 (Palm Oil Research Institute Malaysia, Kuala Lumpur 1990).
Tang, T. L. The possible use of pisifera in plantation. Malay Agric J 48, 57–68 (1971).
Rao, V., Law, I. H., Shaharudin, Z. & Chia, C. C. Ekona and AVROS - a tale of two pisiferas. In 1999 PORIM Int. Palm Oil Congr. (PORIM, Kuala Lumpur 1999).
Sterling, F. & Alvorado, A. Ekona and Calabar as alternative sources of male parents in the commercial production of the oil palm seed. ASD Oil Palm Papers 11, 23–32 (1995).
Xue, Y. et al. Spread of an Inactive Form of Caspase-12 in Humans Is Due to Recent Positive Selection. American Journal of Human Genetics 78, 659–670 (2006).
Kelly, M. & Semsarian, C. Multiple Mutations in Genetic Cardiovascular Disease. Circulation: Cardiovascular Genetics 2, 182 (2009).
Ingles, J. et al. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. Journal of Medical Genetics 42, e59 (2005).
Zheng, X. et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28, 3326–3328 (2012).
Acknowledgements
We would like to acknowledge the contribution of our breeders from Oil Palm Breeding Unit. We also thank Molecular Breeding & Bioinformatics Unit, Sime Darby for sampling of oil palm materials and analytical supports. The study was conducted in Sime Darby Plantation R&D Centre, which is fully supported by a grant from Sime Darby Plantation Division, Malaysia.
Author information
Authors and Affiliations
Contributions
C.T., S.M., D.A. and H.K. conceived and designed the experiment; P.T., S.D.M., P.F., A.O., D.A. and C.T. performed data analysis and interpretations; C.T., S.D.M., P.T., P.F., S.M., F.C. and A.O. drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
This study is fully supported by a grant from Sime Darby Plantation Division, Malaysia. Two authors i.e. F.C. (National University of Singapore, Singapore) and S.M. (University of Nottingham, UK) have received consultancy fees under the same grant. The rest of the authors as the employees to Sime Darby Plantation Division declare that they have no relevant conflicts of interest.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Teh, CK., Muaz, S.D., Tangaya, P. et al. Characterizing haploinsufficiency of SHELL gene to improve fruit form prediction in introgressive hybrids of oil palm. Sci Rep 7, 3118 (2017). https://doi.org/10.1038/s41598-017-03225-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03225-7
This article is cited by
-
A genetic platform for predicting and reducing non-tenera contamination in oil palm (Elaeis guineensis) seed supply
Tree Genetics & Genomes (2021)
-
A practical genome-enabled legitimacy assay for oil palm breeding and seed production
BMC Plant Biology (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.