Abstract
We employed RNA sequencing analysis to reveal dysregulated lncRNAs in lung cancer utilizing 461 lung adenocarcinomas and 156 normal lung tissues from 3 separate cohorts. We found that LINC00152 was highly overexpressed in lung tumors as compared to their adjacent normal tissues. Patients with high LINC00152 expression demonstrate a significantly poorer survival than those with low expression. We verified the diagnostic/prognostic potential of LINC00152 expression in an independent cohort of lung tumor tissues using quantitative RT-PCR. After knockdown of LINC00152 using siRNAs in lung cancer cell lines, both cell proliferation and colony formation were decreased. Cell fractionation and qRT-PCR analysis indicated that LINC00152 is found mainly in the cytoplasm. Treatment with Trichostatin A in cell lines having low LINC00152 expression indicated that histone acetylation may be one mechanism underlying LINC00152 overexpression in NSCLC. Western blot analyses indicated that p38a, STAT1, STAT3, CREB1, CCNE1 and c-MYC proteins were decreased after LINC00152 siRNA treatment. Our study indicates LINC00152 plays an important role in lung tumor growth and is potentially a diagnostic/prognostic marker. Further characterization of LINC00152 in regulating its target proteins may provide a novel therapeutic target of lung cancer.
Similar content being viewed by others
Introduction
It is estimated that 595,690 Americans will die from cancer in 2016, and more than one-quarter of these (158,080) will be due to lung cancer. Lung cancer continues to be the number one cause of cancer related death in both men and women worldwide1. While recent advances using screening CT scans are diagnosing early disease more frequently, the 5-year relative survival is still low (18%). These low survival rates are partly due to the fact that one-half of cases are diagnosed at a higher stage, for which 5-year survival is only 4%1, 2. While multiple molecular events converge to trigger unregulated growth, invasion, and metastasis in lung cancer, the exact mechanisms are not fully understood. Thus there is an urgent need for identification of markers that may aid in the early diagnosis or stratification of lung cancer as well as new therapeutic targets.
Accumulated evidence showed that more than 70% of the human genome is transcribed into primary RNA, but only about 2% encodes for peptide products, with the remainder being noncoding RNAs (ncRNAs)3, 4. These ncRNAs can be divided into two groups based on their transcript lengths: small ncRNAs, which are shorter than 200 bp, and long ncRNAs (lncRNAs), which are longer than 200 bp5. The lncRNAs are usually expressed in a tissue-specific pattern and show a low level of expression and demonstrate low sequence conservation as compared to protein coding RNAs. Through gene expression microarrays and RNA sequencing analysis, hundreds of lncRNAs have been reported to be dysregulated in lung cancer6,7,8; however, only a few have been well characterized regarding their functional role in cancer9. LncRNAs are thought to drive many important cancer phenotypes and disease-related pathways such as controlling cellular proliferation, invasion, development, lineage commitment, immune response, pluripotency and differentiation10,11,12,13. The cellular localization of lncRNAs may determine their function roles, e.g. nuclear lncRNAs are enriched for functions involving chromatin interaction, transcriptional regulation and RNA processing, while cytoplasmic lncRNAs can modulate mRNA stability or translation and influence cellular signaling cascades14.
LINC00152 has been linked to several human cancers and promotes cell proliferation in gastric and hepatocellular carcinoma (HCC)15, 16. Additionally, it may also act as a potential prognostic biomarker and therapeutic target in colorectal cancer, clear cell renal carcinoma and HCC17,18,19. Moreover, since plasma levels of LINC00152 are significantly elevated in patients with gastric cancer, this lncRNA has the potential to serve as a blood-based biomarker for this disease20. The expression of LINC00152 and its functional roles in lung cancer, however, are unexplored. In this study, through analysis of RNA-Seq data from a large cohort of lung cancers, we demonstrate that LINC00152 is highly expressed in lung cancer, and associated with poor patient survival. We validate its expression in an independent cohort of primary lung cancer using RT-PCR and explored its oncogenic functions in lung cancer cell lines, as well as the possible molecular mechanisms involved in lung cancer.
Results
Increased LINC00152 expression is correlated with worse prognosis in patient with lung ADs
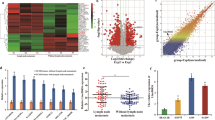
To identify the dysregulated lncRNAs and their diagnostic potential in lung adenocarcinomas (LUAD), the largest subtype of NSCLC, we performed Receiver Operating Characteristic (ROC) curve analysis with combined RNA-Seq data from our cohort (UM 67 LUADs and 6 normal lung tissues), and two other independent published LUAD cohorts, Seo (85 LUADs and 85 normal lung tissues)21 and TCGA (309 LUADs and 73 normal lung tissues)22. We have identified lncRNAs differentially expressed in LUADs as compared to normal lung tissues6. Among the most highly overexpressed lncRNAs, LINC00152 was found to be significantly increased in LUADs. Scatterplots showed that LINC00152 maintained a high expression level in tumors (vs. normal) in the three cohorts (Fig. 1A–C). The area under the curve (AUC) values from ROC analysis were larger than 0.74 in all 3 cohorts (Fig. 1D–F) indicating that LINC00152 may be potentially used as a novel diagnostic marker for this type of lung cancer. We next evaluated the association of LINC00152 and patient survival in two independently published LUAD microarray data sets where survival information was available, Okayama et al. (226 LUADs, stage 1 and 2)23 and TCGA (197 LUADs, stage 1 to 3)22. Kaplan-Meier survival curves and log-rank tests showed that higher expression levels of LINC00152 was significantly correlated with poor patient outcome in the Okayama (p = 0.001) and TCGA data sets (p = 0.03) (Fig. 1G,H), whereas patients with relatively lower levels of LINC00152 expression showed better survival. Taken together, LINC00152 was significantly overexpressed in LUAD across multiple studies and overexpression was a predictor of poor patient survival in LUADs.
Differentially expression of LINC00152 in lung ADs. (A–C) Scatterplots of LINC00152 expression levels in lung AD and normal tissue samples in UM, Seo and TCGA RNA-Seq data sets (y-axis is log2 of FPKM value, LUAD vs. Normal, p < 0.001 in all 3 data sets). (D–F) ROC curves with AUC values of LINC00152 in UM (67 LUAD vs 6 N), Seo (85 LUAD vs 77 N) and TCGA (309 LUAD vs 73 N) RNA-Seq data sets. (G and H) Kaplan-Meier curves and log-rank test of LINC00152 in the Okayama (226 LUADs), and TCGA (197 LUADs) data sets. Higher LINC00152 expression was associated with poor patient survival.
Validation of LINC00152 expression pattern in an independent cohort of LUADs by qRT-PCR
To verify the LINC00152 expression pattern discovered from RNA-Seq and microarray data sets, we performed qRT-PCR using mRNA from an independent cohort from UM including 101 LUADs and 27 normal lung tissues. The results showed that LINC00152 expression levels were significantly higher in LUAD as compared to normal lung tissues (p < 0.001) (Fig. 2A). The AUC was 0.88 indicated LINC00152 expression levels could classify the tumors from normal lung (Fig. 2B). Kaplan-Meier survival curve and the log-rank test indicated that higher expression of LINC00152 was significantly related to worse patient survival (p = 0.003) (Fig. 2C). We also analyzed LINC00152 expression levels and other clinical variables from this validation set, however, no evidence was obtained to support the association between LINC00152 expression and age, gender, smoking, differentiation, tumor stage, lymph node or KRAS mutation status (Supplementary Table 1).
qRT-PCR validation of LINC00152 expression in an independent data set (A–C) and LINC00152 expression in other tumors types (D). (A) Scatterplot indicating LINC00152 expression was higher in tumor (measured by RT-PCR, y-axis is fold change to mean of normal tissues, LUAD vs. normal, p < 0.001 by t test). (B) ROC curve indicated an excellent AUC (0.88) for classifying the 101 LUAD from 27 normal lung tissues based on LINC00152 expression. (C) Kaplan-Meier curve indicated higher LINC00152 expression was unfavorable for patient survival. (D) Comparison of LINC00152 expression in cancers (red) and matched normal (blue) tissues (mean + SEM, log2 of FPKM from RNA-seq data). Lung, bladder, breast, gastric, head and neck, kidney, liver, and thyroid cancers were significantly higher in tumor (vs. normal, t test, p < 0.05). Case number of each cancer is indicated. The original data was downloaded from miTranscriptome with modification.
Overexpression of LINC00152 was reported in colorectal cancer, clear cell renal carcinoma and HCC; however to determine if LINC00152 expression was higher in other cancers, we analyzed RNA-Seq expression data including 6,220 cancers from the MiTranscriptome database24. We found that LINC00152 was increased in most cancers including bladder, breast, gastric, head/neck, kidney, liver, thyroid, cervix, blood, uterus, and colon cancers (FRKM log2 value > 1.4) but not prostate cancer (FRKM log2 = 0.67) (Fig. 2D). Interestingly, we found that squamous cell (LUSC) and large cell (LULC) lung carcinomas also highly expressed LINC00152 (vs. normal). These results indicated that LINC00152 was highly expressed not only in lung cancer but also in other type of cancers and could be potentially useful as a general cancer marker.
LINC00152 overexpression caused by histone acetylation
Gene expression could be influenced by genomic copy number changes, or specific transcription factors coupled with changes in histone and DNA modifications in its gene promoter. We examined DNA copy number changes of the LINC00152 genomic locus by Affymetrix SNP 6.0 arrays on 90 lung adenocarcinomas and 10 normal lung tissues (unpublished data) and did not find any apparent amplification except for small gains in two cases (Fig. 3A). To test whether LINC00152 expression is related to histone acetylation and promoter DNA methylation, we first analyzed its expression levels in 33 lung cancer cell lines from RNA-Seq data. We found that LINC00152 was highly expressed in 30 NSCLC cell lines with lower expression in 3 small cell lung cancer cell lines, H146, H526 and H82 (Fig. 3B). We hypothesized that histone deacetylation or promoter DNA methylation may be the mechanisms causing the low expression of LINC00152 in H146, H526 and H82 cells. We then treated these cell lines with 5-aza-2-deoxycytidine (5-AZA) and/or Trichostatin A (TSA) and found that LINC00152 expression was increased by 16-fold in H526 and 8-fold in H146 cells after TSA treatment, whereas, 5-AZA didn’t change LINC00152 expression (Fig. 3C), indicating that histone acetylation could be one mechanism causing LINC00152 overexpression in NSCLC. We don’t know the reason so far why H146 cells with higher concentrations of TSA (0.5uM) lost the increase in expression of LINC00152. But, we found that these two small cell lung cancers have a different cell growth reaction upon LINC00152 siRNA treatment (We described the results in Supplementary Figure S3).
(A) No amplification of the LINC00152 region was detected as determined by Affymetrix SNP6.0 analysis of 90 lung adenocarcinomas and 10 normal lung tissues. Two tumors with gains were found. (B) LINC00152 maintains a high expression level in most of the non-small cell lung cancer cell lines according to the RNA-Seq data (log2 of FPKM value, p < 0.05), whereas, the three small cell lung cancer cell lines, H526, H146 and H82, have a relative low expression of LINC00152. (C) 5-aza-2-deoxycytidine 5-aza and Trichostatin A (TSA) treatment indicates that deacetylation might result in the low expression of LINC00152 in H146 and H526 cells.
LINC00152 knockdown decreased cell proliferation and colony formation in lung cancer cells
Based on RNA-Seq value in Fig. 3B, all lung cancer cell lines have higher LINC00152 expression level. In order to confirm the expression, we performed RT-PCR for LINC00152 expression on the cells which will be used for proliferation assay. A significant correlation between RNA-Seq and RT-PCR was found using Pearson correlation analysis (Supplementary Figure S1). Small cell lung cancer cell line H526 still is the lowest expression for LINC00152. To minimize the possibility of off-target effects, we used SMARTpool gene specific siRNAs whose knockdown efficiency was greater than 80–90% as determined by qRT-PCR (Fig. 4A and Supplementary Figure S2). To explore the oncogenic function of LINC00152, we examined cell proliferation status after LINC00152 knockdown by siRNA in 12 lung cancer cell lines. A significant decrease in cell proliferation rate (from 18% to 38%) was identified in 9 out of 12 cells measured by WST-1 (Fig. 4B and Supplementary Figure S3A). The cell growth of three cell lines, H1975, H1650 and H146, was not affected by LINC00152 knockdown. PC-9 (EGFR mutant and EGFR tyrosine kinase inhibitor (TKI) sensitive cell line) and H838 (EGFR wild type cell line) cells were the most significantly affected (decreased by 36% and 38%, respectively) and were then chosen for colony formation assay. The number of colonies was markedly reduced after LINC00152 siRNA knockdown in PC-9 and H838 cells (Fig. 4C and D). LINC00152 did not affect cell invasion using Boyden chamber matrix assays in these two lung cancer cell lines (data not shown).
Effects of LINC00152 on cell viability. (A) LINC00152 expression was decreased more than 90% after LINC00152 knockdown with siRNA on PC-9 and H838 cells measured by RT-PCR. (B) Cell proliferation measured by WST-1 assays was conducted on 8 lung cancer cell lines to determine the effect on cell viability after LINC00152 siRNA transfection for 96 hours. Values indicate the mean + SD from three independent experiments. **p < 0.01, *p < 0.05. (C and D) Colony formation was decreased after LINC00152 siRNA transfection on PC-9 and H838 cells. **p < 0.01.
We didn’t find a significant correlation between the LINC00152 expression and the inhibition rate of cell growth by LINC00152 knockdown (Supplementary Figure S3B). The cell growth in H1975 and H1650 cells were not affected by LINC00152 siRNA knockdown although these cells have relative higher level of LINC00152 expression. Whereas, the cell growth in H526 was affected although this cell has relative low level of LINC00152 expression. But the cell growth in H146, another low level of LINC00152 expression cell line, was not affected. This indicates that cell growth will be affected if LINC00152 expression reaches a certain level and the genomic background may be the key factor regarding the role of LINC00152 in cell growth.
Proteins/mRNAs regulated by LINC00152 in lung cancer cells
To provide molecular mechanistic insight into LINC00152 role in regulating lung cancer cell proliferation, we first performed receptor tyrosine kinase (RTK) phosphorylation antibody array analysis, which includes 49 different phosphorylated proteins covering most of the cancer-related pathways. The hosphorylation levels of STAT1 and STAT3 proteins were found to be decreased when LINC00152 was knocked down by siRNA at 72 hours (Supplementary Fig. S4A and B). In order to confirm and identify more altered proteins regulated by LINC00152, we performed Western blot on several cell growth-related proteins. As indicated in Fig. 5A and B, p38α, STAT1, STAT3, CCNE1, CREB1 and c-MYC proteins were decreased after LINC00152 siRNA treatment in PC-9 and H838 cell lines. The total proteins of p38α and CCNE1 in H838 cell line were not changed indicating a different regulation mechanism for p38α and CCNE1 by LINC00152 between PC-9 and H838 cells. We also performed the mRNA expression of these genes (Supplementary Fig. S5), we found that STAT3 mRNA was decreased by 30–40% in both cells, whereas CCNE1 mRNA in H838 was increased by 1.5 fold. Other gene mRNAs (p38a, STAT1, CREB1 and c-MYC) were not changed. These results indicated that LINC00152 regulates STAT3 may be through transcription regulation, whereas other proteins, such as p38a, STAT1, CREB1, and c-MYC may be regulated at the post-transcription. The mechanism of these proteins regulated by LINC00152, such as if through lysosome or ubiquitin dependent process, is warranted to be analyzed in the future.
Proteins regulated by LINC00152 and cellular location of LINC00152. (A) Western blot showing the altered protein expression of p38α, STAT1, STAT3, CREB1, CCNE1, and c-MYC after LINC00152 knockdown with siRNA. (B) Quantitative analysis of image in A using ImageJ software. GAPDH was used as a protein loading control. Each individual band was divided by GAPDH first, then divided by NT. So each protein value (siRNA treated) was relative to their NT (NT = 1). (C and D) qRT-PCR showing the nuclear and cytoplasmic fractions of LINC00152 in PC-9 and H838 cells. GAPDH was used as cytoplasmic control and U1 snRNA as nuclear control. LINC00152 is primarily in cytoplasmic (67.7%). (E) Summary of proteins affected by LINC00152 in this study. As indicated in Fig. 5A and B, all proteins were decreased after LINC00152 knockdown, but CCNE1 and total p38a proteins were not changed in H838 cells.
Since EGFR signaling was reported to be involved in LINC00152 promoting cell proliferation in gastric cancer15, we performed Western blot on EGFR, AKT and ERK1/2 proteins. We found that these proteins were not changed after LINC00152 siRNA treatment at 72 hours (Supplementary Fig. S4C) indicating that EGFR signaling was not involved in LINC00152 regulation in these lung cancer cells.
In order to evaluate the relationship between p38α and STAT3, we performed p38α and STAT3 siRNA knockdown on PC-9 and H838 cells. As shown in Supplementary Fig. S6, the ratios of p-STAT3/t-STAT3P were increased by 1.6–1.7 fold (vs. NT) in both cells after p38a knockdown and the mRNA levels were not changed indicating that p38α may regulate STAT3 at the protein level. Figure 5E summarized the proteins regulated by LINC00152 uncovered in this study. More experiments such as STAT1, CCNE1, CREB1 and c-MYC expression changing after p38a or STAT3 knockdown or overexpression is warranted.
Finally, we examined the cellular localization of LINC00152. RNAs were isolated from total, nuclear and cytoplasmic fractions of these two cell lines. Quantitative RT-PCR indicated that the expression of LINC00152 was mainly located in the cytoplasm in both PC-9 (67.2%) and H838 cells (67.7%) (Fig. 5C and D). LINC00152 expression primarily in cytoplasm may support that LINC00152 plays roles at post-transcriptional level which is different from a recent report showing LINC00152 interaction with EZH2 and transcriptional control of target genes in A549 and SPCA1 cell lines25.
Discussion
As highly tissue-specific drivers of cancer phenotypes, lncRNAs are potentially prime targets for cancer therapy. Many studies have confirmed their utility as biomarkers not only in cancer diagnosis but also for patient prognosis across a variety of cancers26,27,28,29,30.
LINC00152 has been reported to be highly expressed in hepatocellular carcinoma, gastric cancer and clear cell renal carcinoma and is involved in the cancer progression15,16,17,18,19, 25. In the present study, we found that the average levels of LINC00152 in LUAD tissues was significantly higher than those in corresponding normal tissues, and higher expression of LINC00152 was associated with a poor patient survival. These results suggest that LINC00152 may be potentially useful as a marker for lung cancer diagnosis and an indicator of poor survival. We found that knockdown of LINC00152 suppressed tumor cell proliferation and colony formation capability of lung cancer cells but did not affect tumor cell invasion, which was consistent with the results observed in both gastric cancer and hepatocellular carcinomas15, 16.
In different cancer types, and even in different cancer cells, lincRNAs may regulate oncogenesis by different molecular mechanisms. In one study, LINC00152 was found to promote tumor growth through an EGFR-mediated PI3/AKT pathway in gastric cancer cell lines, MGC803 and HGC-2715, whereas in another study with gastric cell lines BGC-823 and SGC-7901 cells, LINC00152 promoted GC tumor cell cycle progression by binding to enhancer of zeste homolog 2 (EZH2), thus silencing the expression of p15 and p2131. In hepatocellular carcinoma, LINC00152 appears to activate the rapamycin (mTOR) pathway by binding to the promoter of EpCAM through cis-regulation, which was confirmed by the Gal4-λN/BoxB reporter system16. In the present study in lung cancer, LINC00152 knockdown leads to reduced several cell growth-related proteins such as STAT3, p38α, CREB1, STAT1, c-MYC and CCNE1. This suggests that the molecular signaling affected by LINC00152 in lung cancer may be different from gastric and liver cancers.
The p38 MAPK pathway is known to regulate gene expression, allowing cells to respond to various extracellular stresses, and different stimuli can consequently activate p38 MAPKs and influence a variety of downstream molecules. CREB1, STAT1 and STAT3 are key factors for p38α signaling32. CREB1 influences cell cycle arrest through increasing expression of CCNE1. The p38 MAPK-regulated-CREB1 pathway was shown to contribute to selenite-induced colorectal cancer cell apoptosis in vitro and in vivo 33. Pretreatment with the p38 inhibitor SB203580, increased expression of p-STAT3 in the lung adenocarcinoma cell line A54934 indicating that STAT3 was downstream of the ERK and p38 signaling pathways. We found that the ratios of p-STAT3/t-STAT3P were increased by 1.6–1.7 fold (vs. NT) in both PC-9 and H838 cells after p38a knockdown and the mRNA levels were not changed indicating that p38α may regulate STAT3 at the protein level.
STAT3 was shown to directly or indirectly upregulate the expression of genes required for uncontrolled proliferation and survival, including the genes that encode c-MYC, cyclin D1 and cyclin D2, BCL-XL, MCL1 and survivin35, 36. The increased expression of STAT1 and STAT3 may also positively affect c-Myc, thus precipitating a series of events during oncogenesis including apoptosis inhibition, cell proliferation, angiogenesis and anti-immune responses. We found that STAT3 mRNA and protein were decreased after LINC00152 knockdown indicated that LINC00152 regulated STAT3 may be at the transcription level. Based on our finding that LINC00152 was mainly located in the cytoplasmic of lung cancer cells, the regulation process targeted to p38α, STAT1, CREB1 and c-MYC growth related proteins may be at the post-transcriptional level such as protein degradation mechanism. The detailed mechanism of these proteins regulated by LINC00152, such as if through lysosome or ubiquitin dependent process, is warranted to be further studied.
Taken together, our study demonstrated that LINC00152 was up-regulated in human LUAD, and was correlated with the poor survival of LUAD patients. Its increased expression of LINC00152 might be involved in lung cancer development, and LINC00152 may serve as a potential marker for diagnosis and prognosis. Further characterization of LINC00152 in regulating STAT3, p38a and other proteins may provide a novel therapeutic target of lung cancer.
Materials and Methods
Cell culture
Human lung cancer cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA). All cell lines were routinely maintained in RPMI 1640 supplemented with 10% FBS. All cell lines were cultured in a humidified incubator in 5% CO2 atmosphere at 37 °C. All cell lines were genotyped for identity at the University of Michigan Sequencing Core and were tested routinely for mycoplasma contamination.
Lung tissue samples
Lung cancer tissues were collected from patients undergoing curative cancer surgery from 1991 to 2013 at the University of Michigan Health System. Informed consent was obtained from all patients with experimental protocols receiving approval from the University of Michigan Institutional Review Board and Ethics Committee. The methods were carried out in accordance with approved guidelines. The specimens collected from surgery were freshly frozen in liquid nitrogen and then stored at −80 °C. Frozen tissues for regions containing a minimum of 70% tumor cellularity, defined by cryostat sectioning, were utilized for RNA isolation. All included patients did not receive preoperative radiation or chemotherapy.
5-aza-2-deoxycytidine and Trichotatin A Treatment
Two cell lines, H526 and H146, showing low expression of LINC00152, were treated with 5-aza-2-deoxycytidine (5-AZA) or Trichotatin A (TSA). H526 and H146 were seeded in 6-well plates at the desired density. After treatment with 5-AZA (2 µM and 5 µM) or TSA (0.1 µM and 1 µM) for 48 hours, RNA from each treatment were collected for LINC00152 expression analysis by qRT-PCR.
siRNA transfection
Cells were seeded in 6-well or 96-well plates at the desired density overnight. The target siRNA oligonucleotides or NT (non-targeting RNA) controls with a final concentration of 10 nM were transfected with lipofectamine RNAiMax Reagent (Invitrogen, USA) in OptiMEM medium according to the manufacturer’s instructions. siRNA knockdown efficiency was confirmed by qRT-PCR.
Cell proliferation and colony formation
All the cell lines were plated in 96-well plates at the desired density. After plating for 24 hours, LINC00152 siRNA and NT control were transfected with Lipofectamine RNAiMax Reagent in OptiMEM medium. After transfection with 10 nM LINC00152 siRNA for 96 hours, cell proliferation reagents (WST-1) (Roche) were added to each well, and the absorbance was measured at wavelengths of 450 nm and 630 nm, according to the manufacturer’s instructions. The cell viability percentages were calculated by normalizing to control siRNA.
Colony formation assay was performed to measure the number of viable cells after LINC00152 knockdown by siRNA. Forty-eight hours after siRNA transfection, PC-9 and H838 cells were trypsinized and counted, and an equal amount of cells (100 or 500 cells/well) were then seeded evenly onto 6-well plates in triplicate. Ten to fourteen days later, colonies were stained with 0.5% crystal violet, and colonies with more than 50 cells were counted.
RNA subcellular isolation and qRT-PCR
To identify the subcellular localization of LINC00152, all the cell lines were plated in 100 mm plates. The cells were harvested at 80–90% confluence. The cytoplasmic and nuclear RNA were extracted following the instructions for the RNA Subcellular Isolation Kit (Active Motif). Adding complete lysis buffer to each cell pellet and centrifuging each sample, cytoplasmic RNA was harvested from the supernatant, while nuclear RNA was present in the pellet. The cDNA was synthesized with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative RT-PCR was performed using Power SYBR Green master Mix (Thermo Fisher Scientific, USA) and performed with an ABI StepOne Real-Time PCR System (Applied Biosystems). Each sample was analyzed in duplicate. The housekeeping gene GAPDH was used as loading controls.
Microarray and RNA sequencing datasets
One published Affymetrix microarray data set representing 226 primary lung AD tissues23 was used for survival analysis. The CEL files of microarray data were normalized using the Robust Multi-array Average (RMA) method37. We also obtained RNA-Seq data sets from Seo21 and TCGA22 consisting of a total of 394 ADs, 212 SCCs and 150 normal lung tissues. Expression levels of transcripts were represented as FPKM38. Our primary outcome was overall survival, censored at five years. The information concerning adjuvant chemotherapy or radiation therapy was obtained from the original papers.
Receptor tyrosine kinases (RTK) signaling antibody array and Western blot analysis
To explore the possible regulation mechanisms of LINC00152, PC-9 and H838 cell lines were seeded on 6-well plates separately at a density of 10,000 cells per well. The cell lysates were collected after LINC00152 siRNA treatment at 72 h. The pathscan RTK antibody array (Cell Signaling) was performed according to the manufacturer’s instructions. Western blots were also performed with 10 µg protein using polyacrylamide gel electrophoresis and transfer to nitrocellulose membranes. After being blocked for 1 h with 5% non-fat milk, the membranes were incubated with primary antibodies on a rolling shaker overnight at 4 °C. After incubation with a secondary antibody for 1 hour at room temperature, the membranes were developed using the ECL kit (Amersham, Arlington Heights, IL) and exposed to X-ray film.
Statistical analysis
Data were analyzed using GraphPad Prism 6 (GraphPad software) and R software. To evaluate the diagnostic potential of LINC00152 in LUAD vs. normal, Receiver Operating Characteristic (ROC) curve analysis was used. It showed the tradeoff between sensitivity and specificity (any increase in sensitivity will be accompanied by a decrease in specificity) for the different possible cut-points of a diagnostic test. The diagnostic accuracy was measured by the AUC (area under the curve). An AUC of 1 represented a perfect test; an AUC of 0.5 represented an imprecise test. The data are presented as the mean ± SEM from triplicate experiments and additional replicates as indicated. The significant differences between groups were calculated with Student’s t-test or paired t-test. Survival analysis was performed using the Kaplan–Meier method, and the curves were compared using the log-rank test. A p value < 0.05 was considered statistically significant.
References
Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2016. CA Cancer J Clin 66, 7–30, doi:10.3322/caac.21332 (2016).
Torre, L. A., Siegel, R. L. & Jemal, A. Lung Cancer Statistics. Adv Exp Med Biol 893, 1–19, doi:10.1007/978-3-319-24223-1_1 (2016).
Haemmerle, M. & Gutschner, T. Long non-coding RNAs in cancer and development: where do we go from here? Int J Mol Sci 16, 1395–1405, doi:ijms16011395 (2015).
Li, C. H. & Chen, Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol 45, 1895–1910, doi:S1357-2725(13)00174-X (2013).
Ma, L., Bajic, V. B. & Zhang, Z. On the classification of long non-coding RNAs. RNA Biol 10, 925–933, doi:24604 (2013).
Wang, L. et al. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 7, 11487–11499, doi:7203 (2016).
White, N. M. et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 15, 429, doi:10.1186/s13059-014-0429-8 (2014).
Xu, G. et al. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One 9, e104044, doi:10.1371/journal.pone.0104044 (2014).
Sang, H., Liu, H., Xiong, P. & Zhu, M. Long non-coding RNA functions in lung cancer. Tumour Biol 36, 4027–4037, doi:10.1007/s13277-015-3449-4 (2015).
Yuan, S. X. et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology 56, 2231–2241, doi:10.1002/hep.25895 (2012).
Hu, X. et al. A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26, 344–357, doi:S1535-6108(14)00300-6 (2014).
Gupta, R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076, doi:nature08975 (2010).
Schmitt, A. M. et al. An inducible long noncoding RNA amplifies DNA damage signaling. Nat Genet 48, 1370–1376, doi:10.1038/ng.3673 (2016).
Batista, P. J. & Chang, H. Y. Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307, doi:10.1016/j.cell.2013.02.012 (2013).
Zhou, J. et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res 34, 135, doi:10.1186/s13046-015-0250-6 (2015).
Ji, J. et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget 6, 42813–42824, doi:5970 (2015).
Qiu, J. J. & Yan, J. B. Long non-coding RNA LINC01296 is a potential prognostic biomarker in patients with colorectal cancer. Tumour Biol 36, 7175–7183, doi:10.1007/s13277-015-3448-5 (2015).
Wu, Y. et al. Long non-coding RNA Linc00152 is a positive prognostic factor for and demonstrates malignant biological behavior in clear cell renal cell carcinoma. Am J Cancer Res 6, 285–299 (2016).
Li, J. et al. HULC and Linc00152 Act as Novel Biomarkers in Predicting Diagnosis of Hepatocellular Carcinoma. Cell Physiol Biochem 37, 687–696, doi:000430387 (2015).
Li, Q. et al. Plasma long noncoding RNA protected by exosomes as a potential stable biomarker for gastric cancer. Tumour Biol 36, 2007–2012, doi:10.1007/s13277-014-2807-y (2015).
Seo, J. S. et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res 22, 2109–2119, doi:gr.145144.112 (2012).
Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550, doi:nature13385 (2014).
Okayama, H. et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res 72, 100–111, doi:0008-5472.CAN-11-1403 (2012).
Iyer, M. K. et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47, 199–208, doi:ng.3192 (2015).
Chen, Q. N. et al. Long intergenic non-coding RNA 00152 promotes lung adenocarcinoma proliferation via interacting with EZH2 and repressing IL24 expression. Mol Cancer 16, 17, doi:10.1186/s12943-017-0581-3 (2017).
Yang, X. J., Huang, C. Q., Peng, C. W., Hou, J. X. & Liu, J. Y. Long noncoding RNA HULC promotes colorectal carcinoma progression through epigenetically repressing NKD2 expression. Gene 592, 172–178, doi:S0378-1119(16)30619-9 (2016).
Guo, W. et al. Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Sci Rep 6, 27931, doi:srep27931 (2016).
Hong, H. H. et al. Long non-coding RNA UCA1 is a predictive biomarker of cancer. Oncotarget, doi:10142 (2016).
Wang, Y. et al. The Long Noncoding RNA MALAT-1 is A Novel Biomarker in Various Cancers: A Meta-analysis Based on the GEO Database and Literature. J Cancer 7, 991–1001, doi:10.7150/jca.14663 jcav07p0991 (2016).
Zhang, S. et al. lncRNA Up-Regulated in Nonmuscle Invasive Bladder Cancer Facilitates Tumor Growth and Acts as a Negative Prognostic Factor of Recurrence. J Urol 196, 1270–1278, doi:S0022-5347(16)30559-6 (2016).
Chen, W. M. et al. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing p15 and p21 in gastric cancer. Oncotarget 7, 9773–9787, doi:6949 (2016).
Cuadrado, A. & Nebreda, A. R. Mechanisms and functions of p38 MAPK signalling. Biochem J 429, 403–417, doi:BJ20100323 (2010).
Hui, K. et al. The p38 MAPK-regulated PKD1/CREB/Bcl-2 pathway contributes to selenite-induced colorectal cancer cell apoptosis in vitro and in vivo. Cancer Lett 354, 189–199, doi:S0304-3835(14)00437-6 (2014).
Xue, P. et al. A novel compound RY10-4 induces apoptosis and inhibits invasion via inhibiting STAT3 through ERK-, p38-dependent pathways in human lung adenocarcinoma A549 cells. Chem Biol Interact 209, 25–34, doi:S0009-2797(13)00316-5 (2014).
Yu, H. & Jove, R. The STATs of cancer–new molecular targets come of age. Nat Rev Cancer 4, 97–105, doi:10.1038/nrc1275 (2004).
Harada, D., Takigawa, N. & Kiura, K. The Role of STAT3 in Non-Small Cell Lung Cancer. Cancers 6, 708–722, doi:10.3390/cancers6020708 (2014).
Irizarry, R. A. et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264, doi:10.1093/biostatistics/4.2.2494/2/249 (2003).
Dhanasekaran, S. M. et al. Transcriptome meta-analysis of lung cancer reveals recurrent aberrations in NRG1 and Hippo pathway genes. Nature communications 5, 5893, doi:10.1038/ncomms6893 (2014).
Acknowledgements
This work was supported in part by the National Institutes of Health (grant R01CA154365 to D.G.B. and A.M.C.; grant U01CA157715 to D.G.B.; grant R21CA205414 to G.C.), the University of Michigan’s Cancer Center Support Grant (P30 CA46592), University of Michigan’s Cancer Center Thoracic Oncology Program Research Grant (G.C.), a University of Michigan Department of Surgery RAC grant (G.C.), and the National Natural Science Foundation of China (NSFC) (81660324 to S.F. and 81360409 to S.B.).
Author information
Authors and Affiliations
Contributions
S.F. performed the major experiments. S.F., D.G. and G.C. designed the study and wrote the manuscript. J.Z., W.S., S.B., L.X. and X.C. provided technical help. G.C. performed statistical analysis. J.L., R.R. and A.C. provided tumor samples and scientific discussion. All authors contributed to the discussion and review of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, S., Zhang, J., Su, W. et al. Overexpression of LINC00152 correlates with poor patient survival and knockdown impairs cell proliferation in lung cancer. Sci Rep 7, 2982 (2017). https://doi.org/10.1038/s41598-017-03043-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03043-x
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.