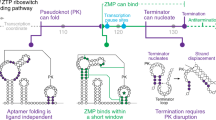
Abstract
Folding of nascent transcripts can be modulated by the RNA polymerase (RNAP) that carries out their transcription, and vice versa. A pause of RNAP during transcription of a preQ1 riboswitch (termed que-PEC) is stabilized by a previously characterized template consensus sequence and the ligand-free conformation of the nascent RNA. Ligand binding to the riboswitch induces RNAP pause release and downstream transcription termination; however, the mechanism by which riboswitch folding modulates pausing is unclear. Here, we report single-particle cryo-electron microscopy reconstructions of que-PEC in ligand-free and ligand-bound states. In the absence of preQ1, the RNA transcript is in an unexpected hyper-translocated state, preventing downstream nucleotide incorporation. Strikingly, on ligand binding, the riboswitch rotates around its helical axis, expanding the surrounding RNAP exit channel and repositioning the transcript for elongation. Our study reveals the tight coupling by which nascent RNA structures and their ligands can functionally regulate the macromolecular transcription machinery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The cryo-EM volumes and maps have been deposited in the Electron Microscopy Data Bank (EMDB) and Protein Database (PDB), respectively. The accession numbers for the cryo-EM density maps reported in this paper are EMD-28845 (RNAP -preQ1 consensus), EMD-29640 (-preQ1 component 0), EMD-29676 (-preQ1 component 1), EMD-29683 (-preQ1 component 2), EMD-29732 (+preQ1 consensus), EMD-29812 (+preQ1 component 0) and EMD-29859 (+preQ1 component 1). The accession numbers for the atomic coordinates reported in this paper are PDB 8F3C (RNAP -preQ1 consensus), PDB 8G0O (-preQ1 component 0), PDB 8G1S(-preQ1 component 1), PDB 8G2W (-preQ1 component 2), PDB 8G4W (+preQ1 consensus), PDB 8GZE (+preQ1 component 0), PDB 8G8Z (+preQ1 component 1). Source data are provided with this paper.
References
Lane, W. J. & Darst, S. A. Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 395, 686–704 (2010).
Bar-Nahum, G. et al. A ratchet mechanism of transcription elongation and its control. Cell 120, 183–193 (2005).
Pan, T. & Sosnick, T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 35, 161–175 (2006).
Chatterjee, S., Chauvier, A., Dandpat, S. S., Artsimovitch, I. & Walter, N. G. A translational riboswitch coordinates nascent transcription–translation coupling. Proc. Natl Acad. Sci. USA 118, e2023426118 (2021).
Artsimovitch, I. & Landick, R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell 109, 193–203 (2002).
Mayer, A. et al. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161, 541–554 (2015).
Gusarov, I. & Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell 3, 495–504 (1999).
Larson, M. H. et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science 344, 1042–1047 (2014).
Kang, J. Y., Mishanina, T. V., Landick, R. & Darst, S. A. Mechanisms of transcriptional pausing in bacteria. J. Mol. Biol. 431, 4007–4029 (2019).
Artsimovitch, I. & Landick, R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl Acad. Sci. USA 97, 7090–7095 (2000).
Guo, X. et al. Structural basis for NusA stabilized transcriptional pausing. Mol. Cell 69, 816–827.e4 (2018).
Abdelkareem, M. et al. Structural basis of transcription: RNA polymerase backtracking and its reactivation. Mol. Cell 75, 298–309.e4 (2019).
Widom, J. R. et al. Ligand modulates cross-coupling between riboswitch folding and transcriptional pausing. Mol. Cell 72, 541–552.e6 (2018).
Sherwood, A. V. & Henkin, T. M. Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374 (2016).
Wang, H. et al. Dual-targeting small-molecule inhibitors of the Staphylococcus aureus FMN riboswitch disrupt riboflavin homeostasis in an infectious setting. Cell Chem. Biol. 24, 576–588.e6 (2017).
Bédard, A.-S. V., Hien, E. D. M. & Lafontaine, D. A. Riboswitch regulation mechanisms: RNA, metabolites and regulatory proteins. Biochim. Biophys. Acta Gene Regul. Mech. 1863, 194501 (2020).
Roth, A. et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 14, 308–317 (2007).
Kang, J. Y. et al. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell 69, 802–815.e1 (2018).
Kang, J. Y. et al. Structural basis of transcription arrest by coliphage HK022 Nun in an Escherichia coli RNA polymerase elongation complex. eLife 6, e25478 (2017).
Webster, M. W. et al. Structural basis of transcription-translation coupling and collision in bacteria. Science 369, 1355–1359 (2020).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Vassylyev, D. G., Vassylyeva, M. N., Perederina, A., Tahirov, T. H. & Artsimovitch, I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature 448, 157–162 (2007).
Nedialkov, Y. A., Nudler, E. & Burton, Z. F. RNA polymerase stalls in a post-translocated register and can hyper-translocate. Transcription 3, 260–269 (2012).
Kent, T., Kashkina, E., Anikin, M. & Temiakov, D. Maintenance of RNA-DNA hybrid length in bacterial RNA polymerases. J. Biol. Chem. 284, 13497–13504 (2009).
Kang, J. Y. et al. Structural basis for transcript elongation control by NusG family universal regulators. Cell 173, 1650–1662.e14 (2018).
Zhu, C. et al. Transcription factors modulate RNA polymerase conformational equilibrium. Nat. Commun. 13, 1546 (2022).
Kang, M., Peterson, R. & Feigon, J. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol. Cell 33, 784–790 (2009).
Toulokhonov, I. & Landick, R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol. Cell 12, 1125–1136 (2003).
Suddala, K. C., Wang, J., Hou, Q. & Walter, N. G. Mg2+ shifts ligand-mediated folding of a riboswitch from induced-fit to conformational selection. J. Am. Chem. Soc. 137, 14075–14083 (2015).
Hein, P. P. et al. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nat. Struct. Mol. Biol. 21, 794–802 (2014).
Landick, R. Transcriptional pausing as a mediator of bacterial gene regulation. Annu. Rev. Microbiol. https://doi.org/10.1146/annurev-micro-051721-043826 (2021).
Zhang, J. & Landick, R. A two-way street: regulatory interplay between RNA polymerase and nascent RNA structure. Trends Biochem. Sci. 41, 293–310 (2016).
Wang, C. et al. Structural basis of transcription-translation coupling. Science 369, 1359–1365 (2020).
Said, N. et al. Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Science 371, eabd1673 (2021).
Yakhnin, A. V. & Babitzke, P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc. Natl Acad. Sci. USA 99, 11067–11072 (2002).
Chauvier, A. et al. Transcriptional pausing at the translation start site operates as a critical checkpoint for riboswitch regulation. Nat. Commun. 8, 13892 (2017).
Komissarova, N. et al. Inhibition of a transcriptional pause by RNA anchoring to RNA polymerase. Mol. Cell 31, 683–694 (2008).
Krupp, F. et al. Structural basis for the action of an all-purpose transcription anti-termination factor. Mol. Cell 74, 143–157.e5 (2019).
Brueckner, F. & Cramer, P. Structural basis of transcription inhibition by α-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 15, 811–818 (2008).
Shu, B. & Gong, P. Structural basis of viral RNA-dependent RNA polymerase catalysis and translocation. Proc. Natl Acad. Sci. USA 113, E4005–E4014 (2016).
Ray-Soni, A., Bellecourt, M. J. & Landick, R. Mechanisms of bacterial transcription termination: all good things must end. Annu. Rev. Biochem. 85, 319–347 (2016).
Shi, J. et al. Structural basis of Q-dependent transcription antitermination. Nat. Commun. 10, 2925 (2019).
Liu, B., Zuo, Y. & Steitz, T. A. Structural basis for transcription reactivation by RapA. Proc. Natl Acad. Sci. USA 112, 2006–2010 (2015).
Heppell, B. et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat. Chem. Biol. 7, 384–392 (2011).
Wickiser, J. K., Winkler, W. C., Breaker, R. R. & Crothers, D. M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Mondal, S., Yakhnin, A. V., Sebastian, A., Albert, I. & Babitzke, P. NusA-dependent transcription termination prevents misregulation of global gene expression. Nat. Microbiol. 1, 15007 (2016).
Chauvier, A., Ajmera, P., Yadav, R. & Walter, N. G. Dynamic competition between a ligand and transcription factor NusA governs riboswitch-mediated transcription regulation. Proc. Natl Acad. Sci. USA 118, e2109026118 (2021).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Carragher, B. et al. Leginon: an automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 132, 33–45 (2000).
Zhang, F. et al. A two-phase improved correlation method for automatic particle selection in cryo-EM. IEEE/ACM Trans. Comput. Biol. Bioinform. 14, 316–325 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Davis, I. W., Murray, L. W., Richardson, J. S. & Richardson, D. C. MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619 (2004).
McGreevy, R., Teo, I., Singharoy, A. & Schulten, K. Advances in the molecular dynamics flexible fitting method for cryo-EM modeling. Methods 100, 50–60 (2016).
Trabuco, L. G., Villa, E., Mitra, K., Frank, J. & Schulten, K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16, 673–683 (2008).
Trabuco, L. G., Villa, E., Schreiner, E., Harrison, C. B. & Schulten, K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods 49, 174–180 (2009).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Denning, E. J., Priyakumar, U. D., Nilsson, L. & Mackerell, A. D. Impact of 2′-hydroxyl sampling on the conformational properties of RNA: update of the CHARMM all-atom additive force field for RNA. J. Comput. Chem. 32, 1929–1943 (2011).
Vanommeslaeghe, K. et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Phillips, J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Landick, R., Wang, D. & Chan, C. L. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 274, 334–353 (1996).
Acknowledgements
We thank J. Widom for the initial design of the que-PEC reconstitution, C. Scull for help with purification of RNAP mutant S1105A, I. Artsimovitch for helpful discussions, UM cryo-EM staff members and UM BSI and LSI for support of the UM cryo-EM facility. This work was supported by National Institutes of Health (NIH) R01 grants GM131922 and GM118524 to N.G.W. and NIH S10OD020011 and S10OD030275 to M.D.O. A portion of the molecular graphics and analyses was performed with UCSF Chimera and ChimeraX developed by the Resource for Biocomputing, Visualization and Informatics at UC San Francisco, with support from NIH P41-GM103311.
Author information
Authors and Affiliations
Contributions
A.C., M.D.O. and N.G.W. conceived the project. A.C., J.C.P. and I.D. devised the methodology. A.C., J.C.P., I.D. and E.E. carried out the investigations. A.C. and J.C.P. wrote the original draft and A.C., J.C.P., I.D., E.E., K.M., M.D.O., A.T.F. and N.G.W. edited and reviewed the article. The work was supervised by A.T.F., M.D.O. and N.G.W. Funding was acquired by A.T.F., M.D.O. and N.G.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Robert Landick, Abhishek Singharoy, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Sara Osman and Beth Moorefield were the primary editors on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Note 1, Figs. 1–10, legends for Videos 1–6 and additional references.
Supplementary Tables
Supplementary Tables 1–3.
Supplementary Video 1
Supplementary video.
Supplementary Video 2
Supplementary video.
Supplementary Video 3
Supplementary video.
Supplementary Video 4
Supplementary video.
Supplementary Video 5
Supplementary video.
Supplementary Video 6
Supplementary video.
Supplementary Data 1
Supplementary data.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chauvier, A., Porta, J.C., Deb, I. et al. Structural basis for control of bacterial RNA polymerase pausing by a riboswitch and its ligand. Nat Struct Mol Biol 30, 902–913 (2023). https://doi.org/10.1038/s41594-023-01002-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-023-01002-x