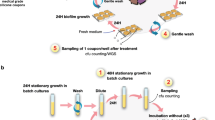
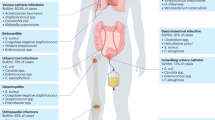
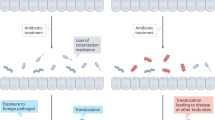
Abstract
Chronic infections caused by microbial biofilms represent an important clinical challenge. The recalcitrance of microbial biofilms to antimicrobials and to the immune system is a major cause of persistence and clinical recurrence of these infections. In this Review, we present the extent of the clinical problem, and the mechanisms underlying the tolerance of biofilms to antibiotics and to host responses. We also explore the role of biofilms in the development of antimicrobial resistance mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Davey, M. E. & O’Toole, G. A. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64, 847–867 (2000).
Rinaudi, L. V. & Giordano, W. An integrated view of biofilm formation in rhizobia. FEMS Microbiol. Lett. 304, 1–11 (2010).
Brandwein, M., Steinberg, D. & Meshner, S. Microbial biofilms and the human skin microbiome. NPJ Biofilms. Microbiomes. 2, 3 (2016).
Hardy, L., Cerca, N., Jespers, V., Vaneechoutte, M. & Crucitti, T. Bacterial biofilms in the vagina. Res. Microbiol. 168, 865–874 (2017).
Motta, J. P., Wallace, J. L., Buret, A. G., Deraison, C. & Vergnolle, N. Gastrointestinal biofilms in health and disease. Nat. Rev. Gastroenterol. Hepatol. 18, 314–334 (2021).
Costerton, J. W. et al. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41, 435–464 (1987).
Bjarnsholt, T. et al. The in vivo biofilm. Trends Microbiol. 21, 466–474 (2013).
Lebeaux, D., Ghigo, J. M. & Beloin, C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543 (2014).
Rupp, M. E. & Karnatak, R. Intravascular catheter-related bloodstream infections. Infect. Dis. Clin. North Am. 32, 765–787 (2018).
Kurtz, S., Ong, K., Lau, E., Mowat, F. & Halpern, M. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J. Bone Jt. Surg. Am. 89, 780–785 (2007).
James, G. A. et al. Biofilms in chronic wounds. Wound Repair. Regen. 16, 37–44 (2008).
Moser, C. et al. Biofilms and host response-helpful or harmful. APMIS 125, 320–338 (2017).
Hoiby, N. et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin. Microbiol. Infect. 21 (Suppl. 1), 1–25 (2015).
Moser, C. et al. Immune responses to Pseudomonas aeruginosa biofilm infections. Front. Immunol. 12, 625597 (2021).
Werdan, K. et al. Mechanisms of infective endocarditis: pathogen-host interaction and risk states. Nat. Rev. Cardiol. 11, 35–50 (2014).
Foster, T. J., Geoghegan, J. A., Ganesh, V. K. & Hook, M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 (2014).
Arciola, C. R., Campoccia, D. & Montanaro, L. Implant infections: adhesion, biofilm formation and immune evasion. Nat. Rev. Microbiol. 16, 397–409 (2018).
Tattevin, P. et al. Risk factors and prognostic impact of left ventricular assist device-associated infections. Am. Heart J. 214, 69–76 (2019).
Trautner, B. W. & Darouiche, R. O. Role of biofilm in catheter-associated urinary tract infection. Am. J. Infect. Control. 32, 177–183 (2004).
Pitts, N. B. et al. Dental caries. Nat. Rev. Dis. Prim. 3, 17030 (2017).
Kimbrell, D. A. & Beutler, B. The evolution and genetics of innate immunity. Nat. Rev. Genet. 2, 256–267 (2001).
Jensen, E. T. et al. Complement activation by Pseudomonas aeruginosa biofilms. Microb. Pathog. 15, 377–388 (1993).
Rybtke, M., Jensen, P. O., Nielsen, C. H. & Tolker-Nielsen, T. The extracellular polysaccharide matrix of pseudomonas aeruginosa biofilms is a determinant of polymorphonuclear leukocyte responses. Infect. Immun. 89, e00631-20 (2020).
Secor, P. R. et al. Pf bacteriophage and their impact on pseudomonas virulence, mammalian immunity, and chronic infections. Front. Immunol. 11, 244 (2020).
Herant, M., Heinrich, V. & Dembo, M. Mechanics of neutrophil phagocytosis: experiments and quantitative models. J. Cell Sci. 119, 1903–1913 (2006).
Alhede, M. et al. Bacterial aggregate size determines phagocytosis efficiency of polymorphonuclear leukocytes. Med. Microbiol. Immunol. 209, 669–680 (2020).
Jones, C. J. & Wozniak, D. J. Psl produced by mucoid pseudomonas aeruginosa contributes to the establishment of biofilms and immune evasion. mBio 8, e00864–17 (2017).
Leid, J. G. et al. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN-gamma-mediated macrophage killing. J. Immunol. 175, 7512–7518 (2005).
de, V. L., Rooijakkers, S. H. M. & van Strijp, J. A. G. Staphylococci evade the innate immune response by disarming neutrophils and forming biofilms. FEBS Lett. 594, 2556–2569 (2020).
Pier, G. B., Coleman, F., Grout, M., Franklin, M. & Ohman, D. E. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69, 1895–1901 (2001).
Bjarnsholt, T. et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol. 44, 547–558 (2009).
Jensen, P. O. et al. Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153, 1329–1338 (2007).
Alhede, M. et al. Pseudomonas aeruginosa recognizes and responds aggressively to the presence of polymorphonuclear leukocytes. Microbiology 155, 3500–3508 (2009).
Mauch, R. M., Jensen, P. O., Moser, C., Levy, C. E. & Hoiby, N. Mechanisms of humoral immune response against Pseudomonas aeruginosa biofilm infection in cystic fibrosis. J. Cyst. Fibros. 17, 143–152 (2018).
Libraty, D. H., Patkar, C. & Torres, B. Staphylococcus aureus reactivation osteomyelitis after 75 years. N. Engl. J. Med. 366, 481–482 (2012).
Brauner, A., Fridman, O., Gefen, O. & Balaban, N. Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016).
Anwar, H., van, B. T., Dasgupta, M., Lam, K. & Costerton, J. W. Interaction of biofilm bacteria with antibiotics in a novel in vitro chemostat system. Antimicrob. Agents Chemother. 33, 1824–1826 (1989).
Hengzhuang, W., Wu, H., Ciofu, O., Song, Z. & Hoiby, N. Pharmacokinetics/pharmacodynamics of colistin and imipenem on mucoid and nonmucoid Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 55, 4469–4474 (2011).
Hoiby, N. et al. Formation of Pseudomonas aeruginosa inhibition zone during tobramycin disk diffusion is due to transition from planktonic to biofilm mode of growth. Int. J. Antimicrob. Agents 53, 564–573 (2019).
Cruz, C. D., Shah, S. & Tammela, P. Defining conditions for biofilm inhibition and eradication assays for Gram-positive clinical reference strains. BMC Microbiol. 18, 173 (2018).
Mottola, C. et al. Susceptibility patterns of Staphylococcus aureus biofilms in diabetic foot infections. BMC Microbiol. 16, 119 (2016).
Macia, M. D., Rojo-Molinero, E. & Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 20, 981–990 (2014).
Fernandez-Barat, L. et al. Phenotypic shift in Pseudomonas aeruginosa populations from cystic fibrosis lungs after 2-week antipseudomonal treatment. J. Cyst. Fibros. 16, 222–229 (2017).
Cao, B. et al. Antibiotic penetration and bacterial killing in a Pseudomonas aeruginosa biofilm model. J. Antimicrob. Chemother. 70, 2057–2063 (2015).
Cao, B. et al. Diffusion retardation by binding of tobramycin in an alginate biofilm model. PLoS ONE 11, e0153616 (2016).
Ciofu, O. & Tolker-Nielsen, T. Tolerance and resistance of pseudomonas aeruginosa biofilms to antimicrobial agents-how p. aeruginosa can escape antibiotics. Front. Microbiol. 10, 913 (2019).
Sweeney, E., Sabnis, A., Edwards, A. M. & Harrison, F. Effect of host-mimicking medium and biofilm growth on the ability of colistin to kill Pseudomonas aeruginosa. Microbiology 166, 1171–1180 (2020).
Bagge, N. et al. Pseudomonas aeruginosa biofilms exposed to imipenem exhibit changes in global gene expression and beta-lactamase and alginate production. Antimicrob. Agents Chemother. 48, 1175–1187 (2004).
Kranjec, C. et al. Staphylococcal biofilms: challenges and novel therapeutic perspectives. Antibiotics 10, 131 (2021).
Ciofu, O., Beveridge, T. J., Kadurugamuwa, J., Walther-Rasmussen, J. & Hoiby, N. Chromosomal beta-lactamase is packaged into membrane vesicles and secreted from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45, 9–13 (2000).
Bagge, N. et al. Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 48, 1168–1174 (2004).
Webb, J. S. et al. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592 (2003).
Christophersen, L. et al. In vivo demonstration of Pseudomonas aeruginosa biofilms as independent pharmacological microcompartments. J. Cyst. Fibros. 19, 996–1003 (2020).
Stewart, P. S. & Franklin, M. J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210 (2008).
Pamp, S. J., Gjermansen, M., Johansen, H. K. & Tolker-Nielsen, T. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 223–240 (2008).
Stokes, J. M., Lopatkin, A. J., Lobritz, M. A. & Collins, J. J. Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259 (2019).
Haagensen, J. et al. Spatiotemporal pharmacodynamics of meropenem- and tobramycin-treated Pseudomonas aeruginosa biofilms. J. Antimicrob. Chemother. 72, 3357–3365 (2017).
Moskowitz, S. M., Foster, J. M., Emerson, J. & Burns, J. L. Clinically feasible biofilm susceptibility assay for isolates of Pseudomonas aeruginosa from patients with cystic fibrosis. J. Clin. Microbiol. 42, 1915–1922 (2004).
Levin, B. R. & Rozen, D. E. Non-inherited antibiotic resistance. Nat. Rev. Microbiol. 4, 556–562 (2006).
Greulich, P., Scott, M., Evans, M. R. & Allen, R. J. Growth-dependent bacterial susceptibility to ribosome-targeting antibiotics. Mol. Syst. Biol. 11, 796 (2015).
Herrmann, G. et al. Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 202, 1585–1592 (2010).
Hansen, C. R., Pressler, T. & Hoiby, N. Early aggressive eradication therapy for intermittent Pseudomonas aeruginosa airway colonization in cystic fibrosis patients: 15 years experience. J. Cyst. Fibros. 7, 523–530 (2008).
Zimmerli, W. & Sendi, P. Role of rifampin against staphylococcal biofilm infections in vitro, in animal models, and in orthopedic-device-related infections. Antimicrob. Agents Chemother. 63, e01746-18 (2019).
Zimmerli, W. & Sendi, P. Orthopaedic biofilm infections. APMIS 125, 353–364 (2017).
Testa, S. et al. Spatial structure affects phage efficacy in infecting dual-strain biofilms of Pseudomonas aeruginosa. Commun. Biol. 2, 405 (2019).
Henriksen, K. et al. P. aeruginosa flow-cell biofilms are enhanced by repeated phage treatments but can be eradicated by phage-ciprofloxacin combination. Pathog. Dis. 77, ftz011 (2019).
Hansen, M. F., Svenningsen, S. L., Roder, H. L., Middelboe, M. & Burmolle, M. Big impact of the tiny: bacteriophage-bacteria interactions in biofilms. Trends Microbiol. 27, 739–752 (2019).
Balaban, N. Q. et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019).
Defraine, V., Fauvart, M. & Michiels, J. Fighting bacterial persistence: current and emerging anti-persister strategies and therapeutics. Drug Resist. Updat. 38, 12–26 (2018).
Fisher, R. A., Gollan, B. & Helaine, S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464 (2017).
Wilmaerts, D., Windels, E. M., Verstraeten, N. & Michiels, J. General mechanisms leading to persister formation and awakening. Trends Genet. 35, 401–411 (2019).
Haussler, S., Tummler, B., Weissbrodt, H., Rohde, M. & Steinmetz, I. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29, 621–625 (1999).
Vulin, C., Leimer, N., Huemer, M., Ackermann, M. & Zinkernagel, A. S. Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters. Nat. Commun. 9, 4074 (2018).
Pestrak, M. J. et al. Pseudomonas aeruginosa rugose small-colony variants evade host clearance, are hyper-inflammatory, and persist in multiple host environments. PLoS Pathog. 14, e1006842 (2018).
Tuchscherr, L., Loffler, B. & Proctor, R. A. Persistence of staphylococcus aureus: multiple metabolic pathways impact the expression of virulence factors in small-colony variants (SCVs). Front. Microbiol. 11, 1028 (2020).
Bogut, A. & Magrys, A. The road to success of coagulase-negative staphylococci: clinical significance of small colony variants and their pathogenic role in persistent infections. Eur. J. Clin. Microbiol. Infect. Dis. 40, 2249–2270 (2021).
Dengler Haunreiter, V. et al. In-host evolution of Staphylococcus epidermidis in a pacemaker-associated endocarditis resulting in increased antibiotic tolerance. Nat. Commun. 10, 1149 (2019).
Wellinghausen, N. et al. Characterization of clinical Enterococcus faecalis small-colony variants. J. Clin. Microbiol. 47, 2802–2811 (2009).
Rani, S. A. et al. Spatial patterns of DNA replication, protein synthesis, and oxygen concentration within bacterial biofilms reveal diverse physiological states. J. Bacteriol. 189, 4223–4233 (2007).
Sonderholm, M. et al. The consequences of being in an infectious biofilm: microenvironmental conditions governing antibiotic tolerance. Int. J. Mol. Sci. 18, 2688 (2017).
Stewart, P. S. et al. Contribution of stress responses to antibiotic tolerance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 59, 3838–3847 (2015).
Kolpen, M. et al. Reinforcement of the bactericidal effect of ciprofloxacin on Pseudomonas aeruginosa biofilm by hyperbaric oxygen treatment. Int. J. Antimicrob. Agents 47, 163–167 (2016).
Tuomanen, E., Cozens, R., Tosch, W., Zak, O. & Tomasz, A. The rate of killing of Escherichia coli by beta-lactam antibiotics is strictly proportional to the rate of bacterial growth. J. Gen. Microbiol. 132, 1297–1304 (1986).
Brochmann, R. P. et al. Bactericidal effect of colistin on planktonic Pseudomonas aeruginosa is independent of hydroxyl radical formation. Int. J. Antimicrob. Agents 43, 140–147 (2014).
Dwyer, D. J. et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl Acad. Sci. USA 111, E2100–E2109 (2014).
Kolpen, M. et al. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax 65, 57–62 (2010).
Worlitzsch, D. et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 109, 317–325 (2002).
Kolpen, M. et al. Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. PLoS ONE 9, e84353 (2014).
Kragh, K. N. et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 82, 4477–4486 (2014).
Kolpen, M. et al. Denitrification by cystic fibrosis pathogens-Stenotrophomonas maltophilia is dormant in sputum. Int. J. Med. Microbiol. 305, 1–10 (2015).
DePas, W. H. et al. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7, e00796–16 (2016).
Chao, Y., Marks, L. R., Pettigrew, M. M. & Hakansson, A. P. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front. Cell Infect. Microbiol. 4, 194 (2014).
Zumft, W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997).
Line, L. et al. Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 5, 554 (2014).
Eschbach, M. et al. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186, 4596–4604 (2004).
Bernier, S. P., Ha, D. G., Khan, W., Merritt, J. H. & O’Toole, G. A. Modulation of Pseudomonas aeruginosa surface-associated group behaviors by individual amino acids through c-di-GMP signaling. Res. Microbiol. 162, 680–688 (2011).
Rossi, E. et al. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 19, 331–342 (2021).
Aanaes, K. et al. Decreased mucosal oxygen tension in the maxillary sinuses in patients with cystic fibrosis. J. Cyst. Fibros. 10, 114–120 (2011).
Aanaes, K. et al. Secretory IgA as a diagnostic tool for Pseudomonas aeruginosa respiratory colonization. J. Cyst. Fibros. 12, 81–87 (2013).
Ciofu, O. et al. P. aeruginosa in the paranasal sinuses and transplanted lungs have similar adaptive mutations as isolates from chronically infected CF lungs. J. Cyst. Fibros. 12, 729–736 (2013).
Schreml, S. et al. Luminescent dual sensors reveal extracellular pH-gradients and hypoxia on chronic wounds that disrupt epidermal repair. Theranostics 4, 721–735 (2014).
James, G. A. et al. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen. 24, 373–383 (2016).
Debats, I. B. et al. Infected chronic wounds show different local and systemic arginine conversion compared with acute wounds. J. Surg. Res. 134, 205–214 (2006).
Fazli, M. et al. Quantitative analysis of the cellular inflammatory response against biofilm bacteria in chronic wounds. Wound Repair Regen. 19, 387–391 (2011).
Trostrup, H. et al. Pseudomonas aeruginosa biofilm aggravates skin inflammatory response in BALB/c mice in a novel chronic wound model. Wound Repair Regen. 21, 292–299 (2013).
Wu, Y., Klapper, I. & Stewart, P. S. Hypoxia arising from concerted oxygen consumption by neutrophils and microorganisms in biofilms. Pathog. Dis. 76, fty043 (2018).
Hunt, T. K., Zederfeldt, B. & Goldstick, T. K. Oxygen and healing. Am. J. Surg. 118, 521–525 (1969).
Frykberg, R. G. et al. A multinational, multicenter, randomized, double-blinded, placebo-controlled trial to evaluate the efficacy of cyclical topical wound oxygen (TWO2) therapy in the treatment of chronic diabetic foot ulcers: the TWO2 Study. Diabetes Care 43, 616–624 (2020).
Boutte, C. C. & Crosson, S. Bacterial lifestyle shapes stringent response activation. Trends Microbiol. 21, 174–180 (2013).
Nguyen, D. et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011).
Hobbs, J. K. & Boraston, A. B. (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect. Dis. 5, 1505–1517 (2019).
Kohanski, M. A., Dwyer, D. J. & Collins, J. J. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435 (2010).
Blazquez, J., Rodriguez-Beltran, J. & Matic, I. Antibiotic-induced genetic variation: how it arises and how it can be prevented. Annu. Rev. Microbiol. 72, 209–230 (2018).
Nang, S. C., Azad, M. A. K., Velkov, T., Zhou, Q. T. & Li, J. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 73, 679–728 (2021).
Fernandez, L. & Hancock, R. E. Adaptive and mutational resistance: role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 25, 661–681 (2012).
Chiang, W. C., Pamp, S. J., Nilsson, M., Givskov, M. & Tolker-Nielsen, T. The metabolically active subpopulation in Pseudomonas aeruginosa biofilms survives exposure to membrane-targeting antimicrobials via distinct molecular mechanisms. FEMS Immunol. Med. Microbiol. 65, 245–256 (2012).
Poole, K. Stress responses as determinants of antimicrobial resistance in Pseudomonas aeruginosa: multidrug efflux and more. Can. J. Microbiol. 60, 783–791 (2014).
Buroni, S. et al. Differential role of RND efflux pumps in antimicrobial drug resistance of sessile and planktonic Burkholderia cenocepacia cells. Antimicrob. Agents Chemother. 58, 7424–7429 (2014).
Zhang, Y. et al. ampG gene of Pseudomonas aeruginosa and its role in beta-lactamase expression. Antimicrob. Agents Chemother. 54, 4772–4779 (2010).
Cornforth, D. M. et al. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl Acad. Sci. USA 115, E5125–E5134 (2018).
Liao, J., Schurr, M. J. & Sauer, K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms. J. Bacteriol. 195, 3352–3363 (2013).
Liao, J. & Sauer, K. The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance. J. Bacteriol. 194, 4823–4836 (2012).
Poudyal, B. & Sauer, K. The PA3177 gene encodes an active diguanylate cyclase that contributes to biofilm antimicrobial tolerance but not biofilm formation by pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62, e01049–18 (2018).
Mah, T. F. et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426, 306–310 (2003).
Beaudoin, T., Zhang, L., Hinz, A. J., Parr, C. J. & Mah, T. F. The biofilm-specific antibiotic resistance gene ndvB is important for expression of ethanol oxidation genes in Pseudomonas aeruginosa biofilms. J. Bacteriol. 194, 3128–3136 (2012).
Fernandez, L., Rodriguez, A. & Garcia, P. Phage or foe: an insight into the impact of viral predation on microbial communities. ISME J. 12, 1171–1179 (2018).
Bjedov, I. et al. Stress-induced mutagenesis in bacteria. Science 300, 1404–1409 (2003).
Conibear, T. C., Collins, S. L. & Webb, J. S. Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS ONE 4, e6289 (2009).
Driffield, K., Miller, K., Bostock, J. M., O’Neill, A. J. & Chopra, I. Increased mutability of Pseudomonas aeruginosa in biofilms. J. Antimicrob. Chemother. 61, 1053–1056 (2008).
Sekowska, A., Wendel, S., Fischer, E. C., Norholm, M. H. H. & Danchin, A. Generation of mutation hotspots in ageing bacterial colonies. Sci. Rep. 6, 2 (2016).
Levin-Reisman, I. et al. Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017).
Levin-Reisman, I., Brauner, A., Ronin, I. & Balaban, N. Q. Epistasis between antibiotic tolerance, persistence, and resistance mutations. Proc. Natl Acad. Sci. USA 116, 14734–14739 (2019).
Melnyk, A. H., Wong, A. & Kassen, R. The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283 (2015).
Rainey, P. B., Buckling, A., Kassen, R. & Travisano, M. The emergence and maintenance of diversity: insights from experimental bacterial populations. Trends Ecol. Evol. 15, 243–247 (2000).
Perron, G. G., Gonzalez, A. & Buckling, A. Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc. Biol. Sci. 274, 2351–2356 (2007).
Santos-Lopez, A., Marshall, C. W., Scribner, M. R., Snyder, D. J. & Cooper, V. S. Evolutionary pathways to antibiotic resistance are dependent upon environmental structure and bacterial lifestyle. eLife 8, e47612 (2019).
Harris, K. B., Flynn, K. M. & Cooper, V. S. Polygenic adaptation and clonal interference enable sustained diversity in experimental Pseudomonas aeruginosa populations. Mol. Biol. Evol. 38, 5359–5375 (2021).
Ahmed, M. N., Porse, A., Sommer, M. O. A., Hoiby, N. & Ciofu, O. Evolution of antibiotic resistance in biofilm and planktonic pseudomonas aeruginosa populations exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 62, e00320-18 (2018).
Ahmed, M. N. et al. Lack of the major multifunctional catalase KatA in pseudomonas aeruginosa accelerates evolution of antibiotic resistance in ciprofloxacin-treated biofilms. Antimicrob. Agents Chemother. 63, e00766-19 (2019).
Ahmed, M. N. et al. The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: beyond selection of antibiotic resistance. NPJ Biofilms. Microbiomes. 6, 28 (2020).
Frimodt-Moller, J. et al. Mutations causing low level antibiotic resistance ensure bacterial survival in antibiotic-treated hosts. Sci. Rep. 8, 12512 (2018).
Ciofu, O., Fussing, V., Bagge, N., Koch, C. & Hoiby, N. Characterization of paired mucoid/non-mucoid Pseudomonas aeruginosa isolates from Danish cystic fibrosis patients: antibiotic resistance, beta-lactamase activity and RiboPrinting. J. Antimicrob. Chemother. 48, 391–396 (2001).
Zaborskyte, G., Andersen, J. B., Kragh, K. N. & Ciofu, O. Real-time monitoring of nfxB mutant occurrence and dynamics in P. aeruginosa biofilm exposed to sub-inhibitory concentrations of ciprofloxacin. Antimicrob. Agents Chemother. 61, e02292-16 (2016).
Scribner, M. R., Santos-Lopez, A., Marshall, C. W., Deitrick, C. & Cooper, V. S. Parallel evolution of tobramycin resistance across species and environments. mBio 11, e00932-20 (2020).
Trampari, E. et al. Exposure of Salmonella biofilms to antibiotic concentrations rapidly selects resistance with collateral tradeoffs. NPJ Biofilms Microbiomes. 7, 3 (2021).
Hausner, M. & Wuertz, S. High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65, 3710–3713 (1999).
Stalder, T. & Top, E. Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms. Microbiomes. 2, 16022 (2016).
Stalder, T. et al. Evolving populations in biofilms contain more persistent plasmids. Mol. Biol. Evol. 37, 1563–1576 (2020).
Savage, V. J., Chopra, I. & O’Neill, A. J. Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 57, 1968–1970 (2013).
Strugeon, E., Tilloy, V., Ploy, M. C. & Da, R. S. The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7, e00868-16 (2016).
Chi, F., Nolte, O., Bergmann, C., Ip, M. & Hakenbeck, R. Crossing the barrier: evolution and spread of a major class of mosaic pbp2x in Streptococcus pneumoniae, S. mitis and S. oralis. Int. J. Med. Microbiol. 297, 503–512 (2007).
Ghigo, J. M. Natural conjugative plasmids induce bacterial biofilm development. Nature 412, 442–445 (2001).
Nolan, L. M. et al. Pseudomonas aeruginosa is capable of natural transformation in biofilms. Microbiology 166, 995–1003 (2020).
Lerminiaux, N. A. & Cameron, A. D. S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 65, 34–44 (2019).
Schooling, S. R. & Beveridge, T. J. Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188, 5945–5957 (2006).
Abe, K., Nomura, N. & Suzuki, S. Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 96, fiaa031 (2020).
Lebeaux, D., Chauhan, A., Rendueles, O. & Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2, 288–356 (2013).
Kaplan, J. B. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 89, 205–218 (2010).
Otto, M. Staphylococcal biofilms. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.GPP3-0023-2018 (2018).
Whiteley, M. et al. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864 (2001).
Valentini, M., Gonzalez, D., Mavridou, D. A. & Filloux, A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr. Opin. Microbiol. 41, 15–20 (2018).
Valentini, M. & Filloux, A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555 (2016).
Hall-Stoodley, L. et al. Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol. Med. Microbiol. 65, 127–145 (2012).
Crabbe, A., Jensen, P. O., Bjarnsholt, T. & Coenye, T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 27, 850–863 (2019).
Acknowledgements
C.M. is supported by Novo Nordisk Foundation (Borregaard Clinical Scientist Fellowship in translational research; grant no. NNF17OC0025074).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Carla Renata Arciola; Mark Webber, who co-reviewed with Eleftheria Trampari; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Glossary
- Tolerance
-
A tolerant population is characterized by a slow rate of killing due to slow growth and low metabolic activity of the bacterial cells, requiring a longer time to kill 99% of the bacterial population than a susceptible population despite the similar values of the minimum inhibitory concentration of antibiotics against both populations.
- Resistance
-
A resistant population is characterized by the lack of killing by antibiotic concentrations above the minimum inhibitory concentration of antibiotics against the susceptible bacterial population.
- Aggregates
-
An assemblage of bacterial cells that can develop into structured biofilms through embedding in a polymeric matrix.
- Lipopolysaccharides
-
Surface molecules, part of the outer membrane of the bacteria with a Gram-negative cell wall.
- Respiratory burst
-
Production of superoxide upon the activation of polymorphonuclear leukocytes with rapid consumption of oxygen.
- Degranulation
-
The liberation of the granular content of antimicrobial agents during the activation of polymorphonuclear leukocytes.
- Pf bacteriophages
-
Temperate filamentous phages that can initiate a chronic infection cycle, during which virions are continuously extruded from the bacterial surface without cell lysis. They can be activated under various conditions such as antibiotic treatment or biofilm growth.
- Rhamnolipids
-
Virulence factors with biosurfactant activity produced by Pseudomonas aeruginosa.
- Persister cells
-
Non-dividing cells that can resume growth and cause relapse of the infection when antibiotic treatment is terminated.
- Bacteriophage
-
Often referred to as simply phages. Naturally occurring viruses that infect bacteria. Lytic phages replicate inside their hosts and release new bacteriophages able to infect more bacteria. Temperate phages incorporate their genetic material into the bacterial chromosome and can be activated under special conditions (for example, treatment with antibiotics).
- Small colony variants
-
(SCVs). Phenotypic variants that are slow-growing and tolerant to immune cells and antimicrobials. They strongly adhere to surfaces and can auto-aggregate.
- Stringent response
-
A universal stress response that is induced by starvation and results in decreased cell growth to promote cell survival.
- SOS response
-
A stress response that counteracts various types of DNA damage.
- RpoS response
-
RpoS is a global stress response regulator induced under various stress conditions dependent on the RpoS alternative sigma factor and a central regulator of many stationary-phase inducible genes. This response enables cells to become more resistant not only to the stress that they first encounter but also to other stress-inducing treatments.
- Membrane vesicles
-
Small (20–400 nm in diameter) lipid bilayer-enclosed particles released from Gram-negative bacteria. They contain both cytoplasmic and periplasmic components. Membrane vesicles from Gram-positive bacteria have also been described.
- Nitrosative stress
-
Stress response to reactive nitrogen species such as nitric oxide (NO−) and its derivatives (nitrous acid (HNO2), peroxinitrite (ONOO–), alkylperoxynitrate (ROONO).
- CRISPR–Cas
-
An immunity mechanism in archaea and bacteria that confers resistance to foreign genetic elements, such as phages and plasmids, and represents an acquired form of immunity.
- Fitness cost
-
The consequence of mutation-induced resistance on the bacterial growth rate.
- Clinical resistance breakpoint
-
Concentrations of antimicrobial that are used to distinguish strains with a high likelihood of treatment success and those in which treatment is more likely to fail.
- Antimicrobial resistance genes
-
(ARGs). Examples of ARGs transmitted by horizontal gene transfer. Conjugation: plasmid-encoded extended spectrum β-lactamases (ESBLs), carbapenemases, plasmid-encoded quinolone resistance genes (qnr), usually on multi-drug resistance plasmids, between different Gram-negative species such as Escherichia coli and Klebsiella pneumoniae, vancomycin-resistance genes between Enterococcus faecalis and Staphylococcus aureus. Transduction: tetracycline and penicillin resistance between S. aureus strains. Natural transformation: resistance to metronidazole in Helicobacter pylori.
- Integrases
-
Enzymes required for the site-specific recombination of integrons. Integrons are genetic elements that facilitate the acquisition and reassembly of gene cassettes encoding products with a variety of functions, including drug resistance.
- Pharmacokinetics and pharmacodynamics
-
Parameters that describe the time-dependent concentration of the antibiotic in the host (pharmacokinetics) and its effect at the infection site (pharmacodynamics).
Rights and permissions
About this article
Cite this article
Ciofu, O., Moser, C., Jensen, P.Ø. et al. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol 20, 621–635 (2022). https://doi.org/10.1038/s41579-022-00682-4
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-022-00682-4
This article is cited by
-
Deciphering antifungal and antibiofilm mechanisms of isobavachalcone against Cryptococcus neoformans through RNA-seq and functional analyses
Microbial Cell Factories (2024)
-
A biodegradable PVA coating constructed on the surface of the implant for preventing bacterial colonization and biofilm formation
Journal of Orthopaedic Surgery and Research (2024)
-
Root volatiles manipulate bacterial biofilms
Nature Ecology & Evolution (2024)
-
Optimization of an in vitro Pseudomonas aeruginosa Biofilm Model to Examine Antibiotic Pharmacodynamics at the Air-Liquid Interface
npj Biofilms and Microbiomes (2024)
-
Tetrahedral framework nucleic acids/hyaluronic acid-methacrylic anhydride hybrid hydrogel with antimicrobial and anti-inflammatory properties for infected wound healing
International Journal of Oral Science (2024)