Abstract
Stochastic optical reconstruction microscopy (STORM) allows wide-field imaging with single-molecule resolution by calculating the coordinates of individual fluorophores from the separation of fluorophore emission in both time and space. Such separation is achieved by photoswitching the fluorophores between a long-lived OFF state and an emissive ON state. Although STORM can image single molecules, molecular counting remains challenging due to undercounting errors from photobleached or overlapping dyes and overcounting artefacts from the repetitive random blinking of dyes. Here we show that fluorophores can be electrochemically switched for STORM imaging (EC-STORM), with excellent control over the switching kinetics, duty cycle and recovery yield. Using EC-STORM, we demonstrate molecular counting by using electrochemical potential to control the photophysics of dyes. The random blinking of dyes is suppressed by a negative potential but the switching-ON event can be activated by a short positive-potential pulse, such that the frequency of ON events scales linearly with the number of underlying dyes. We also demonstrate EC-STORM of tubulin in fixed cells with a spatial resolution as low as ~28 nm and counting of single Alexa 647 fluorophores on various DNA nanoruler structures. This control over fluorophore switching will enable EC-STORM to be broadly applicable in super-resolution imaging and molecular counting.
Similar content being viewed by others
Main
Super-resolution light microscopy methods have revolutionized cell biology by enabling imaging with resolution below the diffraction limit of light such that single proteins and detailed cellular structures can be observed in cells1,2,3,4,5,6. The suite of single-molecule localization microscopy methods, such as stochastic optical reconstruction microscopy (STORM)6,7 and photoactivated localization microscopy4,5 are particularly attractive because they use conventional wide-field approaches but with an order of magnitude higher resolution. This is achieved via the separation of point spread functions (PSFs) of the emission from individual fluorophores and the subsequent localization of emitters. In the STORM technique (direct STORM)6, emitter separation is achieved by photochemically switching the organic dyes into the OFF state using a high-power visible laser. Then, a sparse subset of dyes is switched back to the ON state, either spontaneously or using a second ultraviolet (UV) laser. The locations of the subset of dyes in an imaging frame can then be precisely determined. This process is repeated over many imaging frames and a super-resolution STORM image is reconstructed from the fitted locations. Since a STORM image is built molecule by molecule, it should be possible to determine the number of underlying molecules. However, achieving this can be fairly difficult in practice. For example, the initial switching OFF of the dyes with an intense laser leads to photobleaching8; the bleached molecules will no longer be visible and result in undercounting. Undercounting or missed events can also occur when ON-state fluorophores overlap with densely labelled biological samples. Perhaps more problematic is the random blinking that makes a single dye appear as several molecules, leading to overcounting. This issue is especially difficult to overcome for cyanine-based dyes with prolonged dark time between re-blinking, and temporal grouping will fail9.
The challenges with over- and undercounting of fluorophores are mostly related to control over the switching of fluorophores. To address these challenges, here we propose the use of electrochemistry to switch the fluorophores between their ON and OFF states. The inspiration came from previous research demonstrating that the fluorescence of some fluorophores can be altered by an external electrical potential10. However, in previous research10,11,12,13, only the brightness of the fluorophore was tuned using electrochemical potential with the change synchronized across all the fluorophores. The stochastic switching of molecules between their ON and OFF states, as required for STORM imaging, was not achieved. To understand why the electrochemical switching of fluorophores might be possible requires an understanding of how the fluorophores can be photochemically switched between the ON and OFF states. A simplified Jablonski diagram (Fig. 1a) for the commonly used STORM dye Alexa 647 depicts this mechanism7,14,15. Briefly, the OFF-state transfer of Alexa 647 is a photoinitiated thiol–ene reaction6,16. The majority of Alexa 647 dyes are first excited into a singlet state using a 642 nm laser at high power (10–30 kW cm−2) where they can further intersystem cross into a triplet state17,18. From the triplet state, the fluorophores can then enter a long-lived dark state6 when organothiols react with the polymethine bridge of Alexa 647 and disrupt the conjugated π-electron system16. The reaction is reversible such that the C–S bond for the OFF-state Alexa 647 can dissociate, either by oxygen oxidation or by UV irradiation, which will switch Alexa 647 back to the ON state. In conventional STORM, to keep Alexa 647 in the OFF state for a longer duration, an oxygen scavenger system (like glucose and glucose oxidase) is used to reduce the oxygen concentration by ten times to ~5–20 µM (ref. 19).
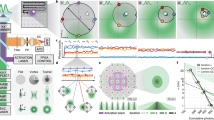
a, Simplified Jablonski diagram showing how Alexa 647 is switched between the ON and OFF states under STORM conditions. In the ON state, Alexa 647 can be excited from the ground state (F0) to the excited singlet state (F1). From F1, Alexa 647 may either relax to the ground state by emitting photons or alternatively undergo intersystem crossing to the dark triplet state (T1). From the triplet state, molecules may return to the ground state via oxidation by oxygen or progress to a long-lived dark state (FH) after reacting with a primary thiol. Alexa 647 molecules in the dark state may return to the ground state via oxidation by oxygen, or alternatively, by exposure to UV radiation. F0 and F1 are called the ON states, whereas T1 and FH are the OFF states. b, Electrochemically switching Alexa 647 ON and OFF. The image frames show that at 0.2 V, the majority of Alexa 647 molecules are in their ON state, whereas at −0.5 V, many of the molecules appear to be in the OFF state. The imaging sample is Alexa 647-labelled microtubules of COS-7 cell, and a single 642 nm laser with an intensity of 2 kW cm−2 was used as the excitation source during imaging. Scale bar, 5 μm. c, Corresponding electrochemical-potential-regulated fluorescence switching curve for the data in b. d, Cyclic voltammograms of bare ITO (green line) and Alexa 647/ITO (black line) in the STORM buffer at a scan rate of 20 mV s−1. e, Correlation between the fluorescence intensity and current change corresponding to the redox peaks at 0.17 V/0.08 V on cyclic voltammograms for the Alexa 647/ITO sample. f, Proposed mechanistic pathways for electrochemically induced switching of Alexa 647. Under a negative potential, the thiol reacts with the –ene functional group at Alexa 647 via an anti-Markovnikov addition to form a thiol ether, and hence, Alexa 647 will be switched into the OFF state. Under a positive potential, C–S-bond dissociation will switch Alexa 647 back to the ON state.
Results
Electrochemical fluorescence switching
To assess whether we could use electrochemistry to control the thiol–ene reaction between thiol and Alexa 647, we used an indium tin oxide (ITO)-coated glass coverslip as the microscope slide to simultaneously perform electrochemistry and fluorescence imaging10,20,21 (Supplementary Fig. 1a). The standard oxygen scavenger, Tris buffer, with cysteamine (a typical STORM buffer) was utilized as the imaging and electrochemical buffer solution. As evident in Fig. 1b,c and Supplementary Video 1, Alexa 647 can be switched between its ON and OFF states by applying positive and negative electrochemical potentials in the STORM buffer, respectively. Much more ON-state Alexa 647 can be observed at a positive potential (0.2 V) than at a negative potential (–0.5 V). Supplementary Video 1 shows that at a constant potential, Alexa 647 molecules exhibit the characteristic stochastic blinking behaviour, whereas the external electrochemical potential has a direct effect on the emitter density.
To understand the underlying electrochemical reactions between the OFF- and ON-state transfer of organic dyes, cyclic voltammetry and differential pulsed voltammetry were performed (Fig. 1d,e and Supplementary Fig. 2b). We noted that two pairs of redox peaks were present when cysteamine was in solution. These two redox peaks could be attributed to the redox reactions of thiol and thiyl radicals22. With Alexa 647 accumulation on ITO, both oxidation and reduction currents increased, and the fluorescence change appropriately correlates with the current change between 0 and 0.3 V, with representative redox peaks at 0.17 V/0.08 V (Fig. 1e and Supplementary Video 2). On the basis of these findings, we proposed a possible radical-involved mechanism (Fig. 1f). When the potential is scanned in the negative direction, several thiol and oxygen reduction reactions may occur, leading to either the formation of thiyl radicals directly or reactive oxygen species23. In the latter case, intermediate oxygen radicals or products, such as the extremely reactive hydroxyl radical (HO•)24, hydroperoxyl radical (HO2•) and hydrogen peroxide (H2O2)25, can react with organothiols to generate thiyl radicals26. In either case, the thiyl radicals can then react with the –ene functional group at a negative potential in the thiol–ene click reaction. When a constant potential was applied to the ITO surface, we observed that the more negative the potential, the more Alexa 647 on the surface was converted into the OFF state. We propose that this is due to the generation of more thiyl radical formations, resulting in more Alexa 647 being switched to the OFF state by thiol addition. Under a positive potential, the oxidation of thiolate anions to disulfides is thermodynamically more favourable, and the electrochemical oxidation of thiols then facilitates the dissociation of C–S (ref. 27). These dissociated thiolate anions rapidly react with other thiols or thiyl radicals to form disulfides instead of propagating with the –ene functional groups on Alexa 647. The electrochemically controlled thiol–ene reaction to switch the fluorescence ON and OFF also works for other STORM dyes, such as ATTO 488 (rhodamine) and ATTO 647N (carbopyronine) dyes (Supplementary Figs. 2 and 3 and Supplementary Videos 3 and 4). The detailed mechanisms of reversible thiol–ene reactions that can be driven by electrochemical potential are, however, not yet completely understood, and further experimental exploration will be necessary.
To study the switching kinetics of Alexa 647 by using EC-STORM, the rate constants for switching the dyes between the fluorescence ON and OFF states (Fig. 2a; kon and koff) using electrochemical switching were measured. Our finding (Fig. 2b) shows that with electrochemical switching, kon can dramatically increase with positive potentials, whereas the more negative potentials increase the rate constant koff. In comparison, using photoswitching (Supplementary Fig. 4), kon increases with the intensity of UV laser and koff increases with the intensity of the 642 nm laser, as others have previously reported6.
a, Measurement for rate constants kon and koff using the electrochemical switching method. By applying a negative potential (–0.80 V) to the ITO surface, Alexa 647 will be switched into the OFF state; a subsequent positive potential (0.15 V) will reversibly switch the Alexa 647 back to the ON state. The rate constant was determined by counting the number of molecules in the field of view as a function of time and then fitting the distribution to a single exponential function. b, Rate constants kon and koff were determined under different positive and negative electrochemical potential values. Note that kon is measured versus –0.80 V, and koff is measured versus 0.15 V. The error bars denote mean ± standard deviation (n = 5 replicates of measurements). c, Box plot for the duty cycle (n = 25 moving windows) at each applied potential. The boxes extend from the 25th to 75th percentiles, with the centre lines representing the median values. The whiskers reach out to the largest and smallest values, not exceeding 1.5 times the interquartile range (IQR) from the upper- and lower-box hinges, respectively. d, Reconstructed STORM images obtained under different conditions using single Alexa 647-labelled DNA-origami-modified ITO slide. The presented images come from different regions of the same slide. A total of 2,000 image frames were collected to reconstruct each STORM image. ‘No switching’ refers to a STORM-style image performed in an oxygen scavenger buffer but without cysteamine such that all the adsorbed Alexa 647 molecules are expected to remain in the ON state. Methods details the settings for recovery yield measurement. Scale bar, 5 µm. e, Corresponding recovered molecules that could be imaged under the ‘no switching’, ‘EC’ and ‘UV’ conditions. The error bars denote mean ± standard deviation (n = 6 replicates of different imaging regions).
As a key switching characteristic that would affect the STORM imaging quality, the duty cycle, defined as the fraction of time a fluorophore is ON within a given period, was studied28. The results shown in Fig. 2c demonstrate that varying the electrochemical potential allows for the tuning of the duty cycle over a wide range, from 4 × 10−5 to 4 × 10−3. In contrast, within the same STORM buffer, and under the same imaging laser power, altering the intensity of the UV laser resulted in duty cycles varying only between 6 × 10−4 and 1 × 10−3. That is, the range of duty-cycle variation achieved through electrochemical control could be 50 times higher than that achieved through UV laser control. The insights into the alterations in duty cycle with varying potentials provide guidance for the application of EC-STORM with different imaging requirements, which will be discussed below.
Successfully recovering the OFF-state fluorophores is critical for resolving the underlying structure. Here we evaluated what portion of fluorophores could be resolved using electrochemical switching. The number of molecules (212 ± 33 in the field of view) was quantified in an oxygen scavenger buffer without cysteamine such that no switching could occur. After adding cysteamine, we performed STORM imaging using both electrochemical and photochemical switching. With the initial OFF switching using an intense 642 nm laser followed by UV activation laser, only ~67% of Alexa 647 was recovered (Fig. 2d,e), with the primary loss occurring during the initial OFF-switching phase (Supplementary Fig. 5). In contrast, with the electrochemical switching OFF and then back ON, 217 ± 39 Alexa 647 molecules were counted, representing a 100% recovery yield. The results indicate that electrochemical switching coupled with a moderate imaging laser is an effective approach for resolving all the underlying fluorophores.
Two- and three-dimensional EC-STORM imaging
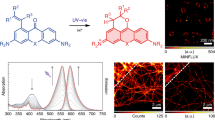
Understanding the electrochemical switching capability of Alexa 647 motivated us to use it for EC-STORM imaging (Fig. 3). A common challenge in conventional STORM imaging on biological samples is the emission overlap of multiple fluorophores. With standard localization algorithms, PSF overlap can lead to false localizations, resulting in blurring of the STORM image, as seen in regions with crossing filaments (Fig. 3b,c). Filtering out the overlapped emitters means the image can appear discontinuous. With the development of ‘multi-emitter fitting’ algorithms29,30,31,32, which greatly improved the localization capability for overlapping PSFs, a compromised resolution can still be obtained32,33. To maintain a low emitter density in conventional STORM, the powers of the coupled lasers during data acquisition8,28 or the imaging buffer and labelling density need to be optimized. With EC-STORM, we demonstrated the capability to adjust the duty cycle with potential, thereby offering effective control over the emitter density (Supplementary Fig. 6a and Supplementary Videos 5 and 6). When a negative potential was applied to the ITO surface, the emitter density could be substantially reduced and less PSF overlapping occurred during EC-STORM imaging compared with the conventional STORM condition (Supplementary Fig. 6b). This reduced the PSF overlapping that led to fewer blurred features in the highlighted regions for EC-STORM compared with conventional STORM (Fig. 3b,c). Using Fourier ring correlation analysis34, we assessed the STORM image resolution. The analysis showed an improvement in the image resolution with EC-STORM compared with STORM in the zoomed-in region, where the tubulins are densely packed (Fig. 3b), with estimated values ranging from ~37 to ~33 nm. A slightly improved resolution was also observed in the low-density region (Fig. 3c), with estimated values from ~30 nm (STORM) to ~28 nm (EC-STORM). The results shown in Fig. 3 demonstrate that applying a negative potential in EC-STORM can lower the duty cycle and reduce the artefacts caused by PSF overlapping.
a, Comparison of STORM and EC-STORM images was conducted for the same region of a cell. The ‘STORM’ condition refers to a 2 kW cm−2 of 642 nm laser and no UV laser was applied; the ‘EC-STORM’ condition refers to a 2 kW cm−2 of 642 nm laser and applying a negative potential from –0.6 to –0.4 V during data acquisition. Scale bar, 5 µm. b,c, Zoomed-in regions highlighted by applying a negative potential; fewer blurred regions at intersecting microtubules were observed in the EC-STORM image. A zoomed-in view of the area highlighted by the yellow square in a (b), and a zoomed-in view of the white square in a (c). Scale bar, 1 µm. Fourier ring correlation curves corresponding to the zoomed-in regions on the left. The imaged COS-7 cells were labelled for microtubules with Alexa 647. The STORM and EC-STORM images were reconstructed from the same number of localization events under each condition.
We also tested the working depth of electrochemically controlled fluorophore switching by imaging Alexa 647-labelled polystyrene beads in highly inclined and laminated optical sheet illumination mode35. This setup allowed us to image Alexa 647 molecules at various z locations (Supplementary Video 7). Our findings confirmed that effective electrochemical switching is applicable for at least 2 µm from the surface (Supplementary Fig. 7a–c). We applied EC-STORM for resolving the Alexa 647-labelled microtubules with a biplane detection approach36 (Supplementary Fig. 7d), and the z-dimension information of the microtubules was obtained. The results confirmed that electrochemical activation is feasible for three-dimensional STORM imaging.
Single-molecule counting
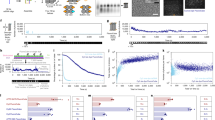
The key capability of EC-STORM that extends beyond imaging is molecular counting. The practical challenges in conventional STORM for molecular counting arise from undercounting issues from the inactivation or photobleaching of fluorophores or the overcounting issue from the random re-blinking of dyes9,37. In Fig. 2, EC-STORM shows exceptional features: it allows for the reversible ON and OFF switching of fluorophores; it has a low duty cycle at a negative potential to avoid PSF overlapping; it can activate 100% of the dyes. Owing to this, we designed a molecular-counting strategy using EC-STORM (Fig. 4a). The electrochemical potential was pulsed between negative and positive values for many cycles. With longer durations at the negative potential, Alexa 647 molecules were held in the OFF state, and only a small subset of molecules would be switched ON during the short positive-potential pulse (Supplementary Video 8). If we assume that the probability of a switching-ON event is identical for each Alexa 647 molecule, the probability of switching-ON events is, therefore, determined by the total underlying number of Alexa 647 molecules. By calibrating the probability of a single fluorophore switching ON at a known site, the number of Alexa 647 molecules in unknown regions can then be calculated on the basis of the ON-event frequency. The results shown in Fig. 4b support our hypothesis that the random blinking of Alexa 647 was efficiently suppressed under a negative potential, and over 95% of the ON events occur at a positive-potential pulse. By employing autocorrelation analysis38, we determined the average ON time of single Alexa 647 molecules from the autocorrelation decay curve (Fig. 4c)39. The result shows that the average ON time of Alexa 647, when subjected to pulsed potential settings, was ~2.5-fold shorter compared with the values observed under the UV activation condition.
a, Schematic of the pulsed electrochemical potential (–0.6 V for 900 ms and 0.1 V for 100 ms for multiple cycles) for switching the fluorophores (i). All the fluorophores could be switched off at –0.6 V, and only a small subset of fluorophores would be briefly switched ON under a short pulse of positive potential. The sample with three fluorophores is assumed to exhibit three times more ON events than a single fluorophore ((ii) and (iii)). b, Number of ON events (blue line) for single Alexa 647-labelled DNA origami in the field of view (25 × 25 µm2) as a function of the pulsed electrochemical potential (red line). c, Autocorrelation analysis of the fluorescent trajectory for single Alexa 647-labelled DNA origami under conventional STORM conditions (blue dashed line; 1 kW cm−2 of 642 nm laser and 10 W cm−2 of UV laser were ON) and EC-STORM where the potential was pulsed (red line; 1 kW cm−2 of 642 nm laser was ON). The average correlation time at 1/e of the maximal autocorrelation amplitude is shown by the dotted line, which was ~323 and ~138 ms for conventional STORM and EC-STORM (pulsed potential), respectively. d, EC-STORM image of a DNA nanoruler acquired by reconstructing 5,000 frames of images as the electrochemical potentials are pulsed. Two ends of the DNA nanoruler were labelled with Alexa 647 and were separated by 94 nm. e, Example for the fluorescent trajectories of two types of DNA nanoruler as the electrochemical potentials are pulsed. A representative schematic of the two types of nanoruler is provided on the left. f, Histograms for the total number of ON events counted on various DNA origami samples. To ensure the exclusion of random re-blinking events, only ON events occurring at a positive potential were counted. A single Alexa 647 refers to the origami labelled with single Alexa 647; nanoruler #1 and #2 refer to the DNA nanorulers illustrated in e with 6 or 12 binding sites for Alexa 647 at each end (total binding sites of 12 or 24), respectively.
As proof of the molecular-counting principle, we then applied EC-STORM to count the DNA nanoruler samples. The DNA nanoruler was designed with Alexa 647 labels at two ends, which were separated by 94 nm. Figure 4d shows a consistent observation with the expected fluorophore separation. A schematic of the two types of DNA nanoruler used for molecular counting is shown in Fig. 4e. The examples for the fluorescence time traces showed that significantly higher number of ON events were observed for nanoruler #2. Figure 4f displays the histograms that highlight the distinctions between the two groups of DNA nanorulers. In contrast, an extensive overlap of the histograms for the total number of ON events between the two nanorulers was observed under the photoactivation condition (Supplementary Fig. 8a). This is probably due to the substantial heterogeneity in Alexa 647 switching under UV activation, with some molecules repeatedly blinking, whereas others are difficult to activate. This indicates that the pulsed potential forces more dyes to enter the ON and OFF switching for more cycles, making single-molecule counting more practical. To assist in quantifying the absolute number of Alexa 647, we applied the DNA origami with one binding site for Alexa 647 for calibration (Fig. 4f). The histogram of the total ON events for a single Alexa 647 shows the peak centred at 1.68 ± 0.10. Using this number, the averaged total number of Alexa 647 molecules at nanoruler #1 and nanoruler #2 were calibrated as 4.10 ± 0.18 and 7.07 ± 0.29, respectively (Table 1).
To gain further evidence for the total number of Alexa 647 molecules bound to each type of nanoruler, we also counted Alexa 647 on the DNA nanoruler from the maximum brightness (Supplementary Fig. 8b–d). In detail, we turned on all the Alexa 647 molecules in the STORM buffer by ramping the potential from –0.8 to 0.6 V. Subsequently, an EC-STORM image was obtained in the same field of view to determine the location for each well-folded DNA nanoruler. The spatial coordinates were used to extract the fluorescence time traces during the ramping of electrochemical potential and estimate the total number of Alexa 647 molecules. This showed consistent numbers of Alexa 647 at nanoruler #1 and nanoruler #2 as obtained from EC-STORM, namely, 4.04 ± 0.10 and 7.24 ± 0.21, respectively (Table 1).
Discussion
The single-molecule localization-based STORM technique demands well-controlled ON and OFF switching of the organic dyes. Here we presented an electrochemical switching method referred to as EC-STORM that affords robust control over switching using Alexa 647 dye as an example. The proposed switching is based on the electrochemically coupled reversible thiol–ene reaction. We thoroughly evaluated the switching kinetics and duty cycles in response to an applied potential.
Our experiments show that the electrochemical switching of fluorophores has several promising benefits for STORM application. First, substituting electrochemical for photochemical switching allows for switching molecules to the OFF state with a moderate imaging laser, and eliminates the need of a UV laser to reactivate the molecules during data acquisition. As such, unwanted photobleaching would be mitigated. A second benefit is the fine control over the duty cycle achieved by altering the potential. This means that the emitter density can be substantially reduced under a negative potential to prevent PSF overlapping, such that densely packed samples can be easily imaged with EC-STORM. A third advantage is that the reversible and fast ON and OFF switching of Alexa 647 using electrochemical potential allows molecular counting. With all the fluorophores switching to the OFF state by a negative potential, the probability of ON events under a positive potential is determined by the underlying number of fluorophores. The working principle for molecular counting using EC-STORM is similar to the quantitative point accumulation for imaging in nanoscale topography (PAINT)40,41, whereas EC-STORM only requires standard dye-labelled antibodies, which reduces the high fluorescence background from floating imager strands, as well as yields faster imaging and counting processes. Finally, the synchronized optical and electrochemical features of the method will allow the study of electrochemical events at the single-molecule level, such as probing electrochemical sensing interfaces and where electrochemistry is occurring within nanoconfined spaces42,43.
Taken together, the electrochemical switching of fluorophores to control their switching characteristics has the potential to transform single-molecule localization microscopy from a powerful imaging tool to single-molecule imaging tools with a broader range of imaging capabilities. Accompanying this dramatic increase in capabilities is the potential simplification of the microscopes to only requiring an imaging laser and an off-the-shelf electrochemical potentiostat.
Methods
Chemicals and materials
Alexa 647-labelled goat anti-rabbit IgG secondary antibody and anti-α-tubulin primary antibody were purchased from Thermo Fisher Scientific. ATTO 647N-labelled goat anti-rabbit IgG secondary antibody was purchased from Sigma-Aldrich. All the solvents used were of the analytical grade unless further indicated. All the chemicals, unless noted otherwise, were of the analytical grade and used as received from Sigma-Aldrich. Aqueous solutions were prepared with Milli-Q water of 18.2 MΩ cm resistivity.
The ITO-coated coverslips (8–12 Ω, 22 × 22 cm2) were purchased from SPI Supplies. Supplementary Fig. 9 provides a detailed comparison between the ITO and glass coverslips, demonstrating similar performance for total internal reflection fluorescence (TIRF) and STORM imaging.
Oxygen-scavenging Tris buffer was prepared as follows: (1) stock buffer A containing 50 mM Tris, 10 mM NaCl (adjusted to pH 8); (2) 10% glucose was added to buffer A followed by adding 0.5 mg ml–1 glucose oxidase and 40 μg ml–1 catalase.
The buffer solutions for conventional STORM and EC-STORM are identical, with 50 mM cysteamine added to the oxygen-scavenging Tris buffer.
Synthesis of single dye-labelled DNA origami
Single-layer, twist-corrected DNA origami rectangles44 were designed with one binding site for an oligonucleotide labelled with the fluorophores (Alexa 647 or ATTO 647N), and eight biotinylated staples for surface attachment. Supplementary Table 1 provides the staple sequences for one binding-site origami. The DNA origami structures were synthesized in a one-pot 60 µl reaction containing 10 nM scaffold strand (M13mp18, Bayou Biolabs), 100 nM staple strands, 200 nM binding site staples and 300 nM dye-labelled oligos (all from Integrated DNA Technologies) in 1× Tris-EDTA buffer (Sigma-Aldrich) with 12.5 mM MgCl2. The solution was annealed using a thermal ramp (65 °C for 15 min and then 60 °C to 44 °C in 160 steps at 6 min per step and then held at 25 °C). DNA origami structures were purified from excess staples and labelled oligos via polyethylene glycol precipitation45. After this, the structures were ready for ITO sample preparation and image acquisition.
The single Alexa 647-labelled DNA origami was utilized for learning the switching rate constants, duty cycle, recovery yield and single-molecule-counting calibration. The single ATTO 647N-labelled DNA origami was used to demonstrate the electrochemical switching of carbopyronine dyes.
DNA-origami-covered ITO sample preparation
Immobilization of DNA origami (GATTAquant) was achieved by modifying clean 22 × 22 cm2 ITO following the below protocols: wash ITO three times with phosphate-buffered saline (PBS) and incubate the ITO with 200 µl of bovine serum albumin–biotin solution (1 mg ml–1 in PBS) for 30 min. Remove the bovine serum albumin–biotin solution and wash it three times with PBS. Make sure not to scratch with the pipette tip on the surface. Incubate the ITO with 200 µl of neutravidin solution (1 mg ml–1 in PBS) for 30 min. Remove the neutravidin solution and wash the chamber three times with 1× PBS supplemented with 10 mM magnesium chloride (the so-called immobilization buffer). Dilute 2 µl of the DNA origami solution with 200 µl immobilization buffer. Incubate the ITO with this solution for 30 min. Wash the ITO three times with the immobilization buffer and store in the immobilization buffer for imaging.
Immunofluorescence staining
Immunostaining was performed using COS-7 cells (ATCC CRL-1651) cultured with Dulbecco’s modified Eagle’s medium fortified with 10% FBS, penicillin and streptomycin, and incubated at 37 °C with 5% CO2. COS-7 cells were plated in six-well plates with ITO slides at ~10,000–20,000 cells per well on the day before fixation. Then, 10 µl of gold nanorods (A12-25-650-PAA-DUG-25, NanoParTz) was added to the ITO coverslip for fiducial drift correction.
The immunostaining procedure for microtubules consisted of primary fixation and permeabilization for 90 s with 0.40% glutaraldehyde (Sigma) and 0.25% Triton X-100 (Sigma) in PBS; second fixation for 7 min at 37 °C with 4.00% glutaraldehyde in PBS, washing with PBS, quenching with 0.40% NaBH4 (Sigma) for 10 min, washing with PBS, incubation for 3 h with rabbit anti-α-tubulin monoclonal antibody (ab216650, Abcam) diluted to 2 µg ml−1 in a blocking buffer (0.20% Triton X-100 and 3.00% BSA in PBS), washing with PBS, incubation for 1 h with secondary anti-rabbit IgG antibodies labelled with Alexa 647 or ATTO 647N at a concentration of ~2.5 µg ml−1 in PBS and washing with PBS; third fixation for 5 min with 2.00% paraformaldehyde in PBS; and finally washing with PBS. Note that at the current stage, EC-STORM is applicable only to fixed cells, and a cell permeabilization step is essential. This step not only allows the entry of antibodies during immunostaining but also enables the effectiveness of electrochemical potential within the cell.
Electrochemistry
Cyclic voltammetry, chronoamperometry and differential pulsed voltammetry were performed by the SP-200 potentiostat (Bio-Logic). All the electrochemistry measurements were carried out in a custom chamber (Chamlide EC 22, Live Cell Instrument) containing an Ag|AgCl|3 M KCl reference electrode and a Pt-wire counter-electrode, where the working electrode was the ITO-coated coverslips.
STORM imaging and analysis
The TIRF images were collected on a ZEISS Elyra SP2 super-resolution photoactivated localization microscope. The collimated and linearly p-polarized 642 nm laser was reflected from the 642 nm long-pass dichroic mirror and focused at the back focal plane of the ×100, 1.46-numerical-aperture oil objective. The focus is laterally shifted along the back focal plane to provide either episcopic illumination (0°) or TIRF illumination (66.7°). The TIRF angle was identical between the glass and ITO surface. The fluorescence was collected by the same objective and guided to a cooled electron-multiplying charge-coupled device camera (iXon DU-897). For STORM imaging, 1–4 kW cm−2 of 642 nm laser was used for excitation with less than 50 W cm−2 of 405 nm laser for photoactivation. For electrochemistry-based STORM, only 1–4 kW cm−2 of 642 nm laser is used. For two-dimensional STORM, 2,000–20,000 images were acquired on a single electron-multiplying charge-coupled device camera with an exposure time of 33–100 ms and camera gain of 80–120. The localization analysis was performed in Zen 2.3 software (black version). Cross-correlation-based drift correction provided in ZEISS Zen software was used for sample drift correction.
For the STORM imaging of the microtubule samples, an initial OFF-switching step is needed to turn off the ON-state fluorophores. The practice is that we set the light path in the episcopic-illumination mode. For conventional STORM, a 5 kW cm−2 of 642 nm laser is turned on for 30 s, and all the fluorophores in the focusing region enter into the blinking mode. In EC-STORM, a negative potential (–0.8 V) is applied to the surface as a moderate-power 642 nm laser (2 kW cm−2) is turned on for 5–10 s; the fluorophores in the focusing region are quickly turned OFF. Subsequently, we set the light path to the TIRF mode for conventional STORM or EC-STORM imaging.
A biplane detection approach was applied to reconstruct the three-dimensional STORM image under the electrochemical switching mode. The fluorescence signal from Alexa 647 was evenly split onto two cameras by a long-pass dichroic beamsplitter (690 nm, AHF Analysetechnik). The tube lens in the shorter-wavelength light path was shifted closer to the camera so that the molecules located at 600 nm higher in the sample space are focused onto this camera. Before three-dimensional image acquisition, the sample with 100 nm microspheres (orange fluorescence (540/560 nm), Thermo Fisher Scientific) was imaged in the TIRF mode for two camera alignments. The beads were also used for axial-distance calibration, where the piezo-stage was moved up at 50 nm step size from –1,350 to +1,350 nm relative to the middle point of the two focal planes as the images were collected. During imaging, the objective was parked in the middle of the two focal planes to produce maximal contrast in the shape variation of the PSF across the two cameras. To reconstruct the three-dimensional EC-STORM image, 30,000 images were simultaneously acquired on two separate cameras with an exposure time of 33 ms. The exposure time of the two cameras was synchronized by the internal trigger provided in ZEISS Elyra.
Rate constant for Alexa 647 switching
To measure the rate constants kon and koff using the electrochemical switching method, the Alexa 647-labelled tubulin samples were imaged under a 642 nm reading laser (2 kW cm−2). By applying a negative potential to the ITO surface, Alexa 647 molecules are switched into the OFF state; a subsequent positive potential reversibly switches them back to the ON state. The rate constant was determined by counting the number of molecules in the field of view as a function of time and then fitting the distribution to a single exponential function (Fig. 2a). For EC-STORM, the rate constant kon at each potential was measured versus –0.80 V and koff at each potential was measured versus 0.15 V. For comparison, the rate constants kon and koff using photochemical switching were measured by irradiating the sample with a strong 642 nm laser to switch OFF Alexa 647 (koff) first and then reduce the 642 nm laser to 2 kW cm−2 on and turn on the UV light to switch the OFF-state fluorophores back to the ON state (kon).
Duty cycle
The single Alexa 647-labelled DNA origami sample was utilized for measuring the duty cycle (fraction of time a molecule spends in its fluorescence-ON state28,46). The measurement process always commenced in a new region of interest, where the Alexa 647 molecules had not switched to the OFF state. These initial ON frames were utilized to determine the location of each DNA origami. Once the molecules entered the stochastic blinking mode, single-molecule fluorescence time traces were extracted. Within a measurement time window of 300 s, the last fluorescence-ON time was assumed to indicate photobleaching of the molecule. The survival fraction was dynamically calculated in a moving window of 5 s as the fraction of molecules that had not undergone photobleaching. The duty cycle was dynamically calculated in a moving window of 15 s. As the molecules experienced photobleaching over time, the duty cycle was normalized by the fraction of molecules that survived in the moving window.
Recovery yield comparison
The STORM images were obtained under different conditions for the same single Alexa 647 molecule-covered ITO sample, but in different regions. Here 2,000 frames were collected to reconstruct the final STORM image. ‘No switching’ represents the STORM image in the oxygen scavenger buffer without cysteamine in which all the Alexa 647 molecules were in the ON state. Under the ‘EC’ condition, a –0.8 V potential was applied for 10 s for switching off the dyes and then an electrochemical potential was applied from –0.10 to 0.35 V during data collection to ensure a higher number of Alexa 647 molecules could be activated to the ON state. Under the ‘UV’ condition, a 10 kW cm−2 of 642 nm laser was applied for 10 s for the initial switching OFF of the dyes, and the UV laser intensity was increased from 10 to 200 W cm−2 during data collection. Then, 1 kW cm−2 of 642 nm laser was turned on during data collection for all the conditions. Under the EC and UV conditions, after 500 frames, there are very few molecules to make sure we have imaged nearly all the molecules that have been turned ON.
Molecular counting using pulsed electrochemical potential (EC-STORM)
The electrochemical potential was pulsed between –0.6 V for 900 ms and 0.1 V for 100 ms for 250 cycles. A STORM image was generated as the potential was pulsing. To filter out the complete nanorulers with two labelling sites, we first examined the pairwise Euclidean distance between all the possible pairs of localization events in the STORM table. Localization events that belong to the same nanoruler tend to be more closely packed in space; therefore, the pairwise distance between the localization events of the same nanoruler was much smaller than the distance between the different nanorulers. Once the densely packed, nanoruler-like regions were isolated, we analysed the distance between the localization events inside the region of interest to its centre of mass. A single-spotted nanoruler due to misfolding-related damage would produce a much smaller value compared with a dual-spotted nanoruler and hence was excluded from the STORM image. Once the dual-spotted nanoruler was identified, the localization events within the nanoruler were segregated into two groups on the basis of the pairwise molecule distance. The distance between these two groups (from centre to another centre of the group) will be analysed (Fig. 4d). From the constructed STORM image, the localization for each DNA origami is obtained; we then analyse the fluorescence trajectories of these well-folded DNA nanorulers from the raw videos and count the total number of ON events.
Absolute molecular counting at the DNA nanoruler using maximum brightness
First, we aimed to activate all the Alexa 647 molecules on the DNA nanoruler by ramping the electrochemical potential from –0.8 to 0.6 V and deactivate the dyes by reversing the potential to –0.8 V. To enhance the statistical accuracy of molecular counting, we repeated this potential-ramping process for three cycles and recorded the first video. Second, to identify the locations of well-folded and labelled DNA nanorulers, we performed EC-STORM in the same region. We took a second video comprising 5,000 frames with the applied potential linearly increasing from –0.2 to 0.2 V. The locations of intact nanorulers, with the two ends separated by 94 nm, were obtained from the reconstructed EC-STORM image. We excluded the degraded DNA nanorulers that exhibited only one end. Third, using the locations of the intact DNA nanorulers, we extracted the fluorescence trajectories from the first video for each DNA nanoruler. By grouping fluorescent events with similar intensity in the intensity–time trace and plotting them in descending order, we generated a stepwise fluorescence decay curve (Supplementary Fig. 9c). From this curve, we estimated the fluorescence intensity of single dyes. We then determined the total number of Alexa 647 molecules at each DNA nanoruler on the basis of maximum brightness.
Data availability
Source data are available via Dryad at https://doi.org/10.5061/dryad.7pvmcvdx9 (ref. 47). Some of the raw data for EC-STORM or conventional STORM are not suitable for distribution through public repositories due to the large file size and are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
Custom MATLAB (MathWorks) codes used for data processing are available via Dryad at https://doi.org/10.5061/dryad.7pvmcvdx9 (ref. 47).
References
Dertinger, T., Colyer, R., Iyer, G., Weiss, S. & Enderlein, J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI). Proc. Natl Acad. Sci. USA 106, 22287–22292 (2009).
Betzig, E. Proposed method for molecular optical imaging. Opt. Lett. 20, 237–239 (1995).
Hell, S. W. & Wichmann, J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780–782 (1994).
Hess, S. T., Girirajan, T. P. & Mason, M. D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. 47, 6172–6176 (2008).
Rust, M. J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006).
Diekmann, R. et al. Optimizing imaging speed and excitation intensity for single-molecule localization microscopy. Nat. Methods 17, 909–912 (2020).
Zhao, Z. W. et al. Spatial organization of RNA polymerase II inside a mammalian cell nucleus revealed by reflected light-sheet superresolution microscopy. Proc. Natl Acad. Sci. USA 111, 681–686 (2014).
Fan, S. et al. Observing the reversible single molecule electrochemistry of Alexa Fluor 647 dyes by total internal reflection fluorescence microscopy. Angew. Chem. Int. Ed. 131, 14637–14640 (2019).
Hilczer, M., Traytak, S. & Tachiya, M. Electric field effects on fluorescence quenching due to electron transfer. J. Chem. Phys. 115, 11249–11253 (2001).
Guille‐Collignon, M., Delacotte, J., Lemaître, F., Labbé, E. & Buriez, O. Electrochemical fluorescence switch of organic fluorescent or fluorogenic molecules. Chem. Rec. 21, 2193–2202 (2021).
Yang, Y. et al. Electrochemical fluorescence switching of enhanced green fluorescent protein. Biosens. Bioelectron. 237, 115467 (2023).
Li, H. & Vaughan, J. C. Switchable fluorophores for single-molecule localization microscopy. Chem. Rev. 118, 9412–9454 (2018).
Bates, M., Huang, B., Dempsey, G. T. & Zhuang, X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007).
Van de Linde, S. & Sauer, M. How to switch a fluorophore: from undesired blinking to controlled photoswitching. Chem. Soc. Rev. 43, 1076–1087 (2014).
Gidi, Y. et al. Unifying mechanism for thiol-induced photoswitching and photostability of cyanine dyes. J. Am. Chem. Soc. 142, 12681–12689 (2020).
Dempsey, G. T. et al. Photoswitching mechanism of cyanine dyes. J. Am. Chem. Soc. 131, 18192–18193 (2009).
Baumann, R. P., Penketh, P. G., Seow, H. A., Shyam, K. & Sartorelli, A. C. Generation of oxygen deficiency in cell culture using a two-enzyme system to evaluate agents targeting hypoxic tumor cells. Radiat. Res. 170, 651–660 (2008).
Lu, X. et al. Monolayer surface chemistry enables 2-colour single molecule localisation microscopy of adhesive ligands and adhesion proteins. Nat. Commun. 9, 3320 (2018).
Ma, Y., Macmillan, A., Yang, Y. & Gaus, K. Lifetime based axial contrast enable simple 3D-STED imaging. Methods. Appl. Fluoresc. 10, 035001 (2022).
Singh, P. et al. Electrochemical capture and release of carbon dioxide using a disulfide–thiocarbonate redox cycle. J. Am. Chem. Soc. 139, 1033–1036 (2017).
Anderson, L. C., Xu, M., Mooney, C. E., Rosynek, M. P. & Lunsford, J. H. Hydroxyl radical formation during the reaction of oxygen with methane or water over basic lanthanide oxide catalysts. J. Am. Chem. Soc. 115, 6322–6326 (1993).
Anbar, M., Meyerstein, D. & Neta, P. The reactivity of aromatic compounds toward hydroxyl radicals. J. Phys. Chem. 70, 2660–2662 (1966).
Nordberg, J. & Arnér, E. S. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31, 1287–1312 (2001).
Fava, A., Reichenbach, G. & Peron, U. Kinetics of the thiol-disulfide exchange. II. Oxygen-promoted free-radical exchange between aromatic thiols and disulfides. J. Am. Chem. Soc. 89, 6696–6700 (1967).
Antonello, S., Benassi, R., Gavioli, G., Taddei, F. & Maran, F. Theoretical and electrochemical analysis of dissociative electron transfers proceeding through formation of loose radical anion species: reduction of symmetrical and unsymmetrical disulfides. J. Am. Chem. Soc. 124, 7529–7538 (2002).
Dempsey, G. T., Vaughan, J. C., Chen, K. H., Bates, M. & Zhuang, X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods 8, 1027–1036 (2011).
Sage, D. et al. Super-resolution fight club: assessment of 2D and 3D single-molecule localization microscopy software. Nat. Methods 16, 387–395 (2019).
Marsh, R. J. et al. Artifact-free high-density localization microscopy analysis. Nat. Methods 15, 689–692 (2018).
Mailfert, S. et al. A theoretical high-density nanoscopy study leads to the design of UNLOC, a parameter-free algorithm. Biophys. J. 115, 565–576 (2018).
Zhu, L., Zhang, W., Elnatan, D. & Huang, B. Faster STORM using compressed sensing. Nat. Methods 9, 721–723 (2012).
Ram, S., Ward, E. S. & Ober, R. J. Beyond Rayleigh’s criterion: a resolution measure with application to single-molecule microscopy. Proc. Natl Acad. Sci. USA 103, 4457–4462 (2006).
Banterle, N., Bui, K. H., Lemke, E. A. & Beck, M. Fourier ring correlation as a resolution criterion for super-resolution microscopy. J. Struct. Biol. 183, 363–367 (2013).
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008).
Juette, M. F. et al. Three-dimensional sub–100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529 (2008).
Annibale, P., Vanni, S., Scarselli, M., Rothlisberger, U. & Radenovic, A. Identification of clustering artifacts in photoactivated localization microscopy. Nat. Methods 8, 527–528 (2011).
Basché, T., Moerner, W., Orrit, M. & Wild, U. Single-Molecule Optical Detection, Imaging and Spectroscopy (John Wiley & Sons, 2008).
Dickson, R. M., Cubitt, A. B., Tsien, R. Y. & Moerner, W. E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 388, 355–358 (1997).
Jungmann, R. et al. Quantitative super-resolution imaging with qPAINT. Nat. Methods 13, 439–442 (2016).
Strauss, S. et al. Modified aptamers enable quantitative sub-10-nm cellular DNA-PAINT imaging. Nat. Methods 15, 685–688 (2018).
Liu, S.-C. et al. An advanced optical–electrochemical nanopore measurement system for single-molecule analysis. Rev. Sci. Instrum. 92, 121301 (2021).
Xue, L. et al. Solid-state nanopore sensors. Nat. Rev. Mater. 5, 931–951 (2020).
Schnitzbauer, J., Strauss, M. T., Schlichthaerle, T., Schueder, F. & Jungmann, R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 12, 1198–1228 (2017).
Stahl, E., Martin, T. G., Praetorius, F. & Dietz, H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. 126, 12949–12954 (2014).
Huang, B., Babcock, H. & Zhuang, X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143, 1047–1058 (2010).
Yang, Y. et al. Source data for ‘Electrochemically controlled blinking of fluorophores for quantitative STORM imaging’. Dryad https://doi.org/10.5061/dryad.7pvmcvdx9 (2024).
Acknowledgements
We thank S. Ciampi and T. Boecking for comments and discussions, and G. Ball for sharing insights into the experimental system. We acknowledge the technical assistance from the Katharina Gaus Light Microscopy Facility, University of New South Wales. J.J.G. acknowledges the Australian Research Council Discovery Grant Program (DP220103024) and a National Health and Medical Research Council Investigator Grant (GNT1196648). Y.M. acknowledges the Earlier Career Fellowship funding from the National Health and Medical Research Council of Australia (APP1139003).
Funding
Open access funding provided through UNSW Library.
Author information
Authors and Affiliations
Contributions
J.J.G. and K.G. designed the project. Y.Y. and Y.M. conceived the study, designed and performed the experiments, analysed the data and wrote the paper. Y.M. wrote the custom code in MATLAB R2023b for analysing the data. J.F.B. and L.K.L. synthesized and labelled the single-site DNA origami. R.D.T. helped revise the paper. J.J.G. supervised all the work and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Photonics thanks Christian Eggeling and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–9 and discussion.
Supplementary Video 1
Electrochemical-potential-modulated fluorescence switching of Alexa 647 in oxygen scavenger Tris buffer with 50 mM cysteamine.
Supplementary Video 2
The ON-state Alexa 647 molecule density change during cyclic voltammetry scan.
Supplementary Video 3
The TIRF image series for ATTO 488 switching under ‘STORM’ and ‘EC-STORM’ conditions. The ‘STORM’ condition refers to 2 kW cm−2 of 642 nm laser and 100 W cm−2 of UV laser was switched on for 1 s and then off for 2 s; the ‘EC-STORM’ condition refers to 2 kW cm−2 of 642 nm laser and applying a pulsed potential between –0.5 V (2 s) and 0.1 V (1 s).
Supplementary Video 4
Electrochemical-potential-modulated fluorescence switching of ATTO 647N in STORM buffer.
Supplementary Video 5
The ON-state Alexa 647 molecule density when the electrochemical potentials are switched between several relative positive values (–0.4, –0.2, –0.1, 0 and 0.1 V) and –0.7 V for 10 s at each still potential.
Supplementary Video 6
The ON-state Alexa 647 molecule density at different positive potentials as indicated.
Supplementary Video 7
When Alexa 647-labelled polystyrene beads (diameter, 100 µm) are placed on an ITO electrode in STORM buffer, the Alexa 647 on the beads shows the switching response to the electrochemical potential. The electrochemical potential was switched between 0.1 V for 0.2 s and –0.8 V for 0.5 s for multiple cycles. The video was recorded under the highly inclined and laminated optical sheet illumination mode, with a frame rate of 50 ms.
Supplementary Video 8
The ON and OFF switching of single Alexa 647-labelled DNA origami sample under EC-STORM where the potential was pulsed between –0.6 V for 900 ms and 0.1 V for 100 ms for multiple cycles.
Supplementary Table 1
Information with all the staple sequences for synthesis of one-binding site origami.
Source data
Source Data Fig. 1
Raw cyclic voltammetry data and raw fluorescence intensity data.
Source Data Fig. 2
Statistical source data and raw STORM table exported from ZEN software.
Source Data Fig. 3
Raw STORM table exported from ZEN software (1–9,999 lines only).
Source Data Fig. 4
Statistical source data and raw STORM table exported from ZEN software.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Ma, Y., Berengut, J.F. et al. Electrochemically controlled blinking of fluorophores for quantitative STORM imaging. Nat. Photon. (2024). https://doi.org/10.1038/s41566-024-01431-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41566-024-01431-0