Abstract
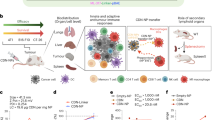
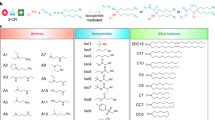
The efficacy of STING (stimulator of interferon genes) agonists is due to various factors, primarily inefficient intracellular delivery, low/lack of endogenous STING expression in many tumours, and a complex balance between tumour control and progression. Here we report a universal STING mimic (uniSTING) based on a polymeric architecture. UniSTING activates STING signalling in a range of mouse and human cell types, independent of endogenous STING expression, and selectively stimulates tumour control IRF3/IFN-I pathways, but not tumour progression NF-κB pathways. Intratumoural or systemic injection of uniSTING-mRNA via lipid nanoparticles (LNPs) results in potent antitumour efficacy across established and advanced metastatic tumour models, including triple-negative breast cancer, lung cancer, melanoma and orthotopic/metastatic liver malignancies. Furthermore, uniSTING displays an effective antitumour response superior to 2′3′-cGAMP and ADU-S100. By favouring IRF3/IFN-I activity over the proinflammatory NF-κB signalling pathway, uniSTING promotes dendritic cell maturation and antigen-specific CD8+ T-cell responses. Extracellular vesicles released from uniSTING-treated tumour cells further sensitize dendritic cells via exosome-containing miRNAs that reduced the immunosuppressive Wnt2b, and a combination of LNP-uniSTING-mRNA with α-Wnt2b antibodies synergistically inhibits tumour growth and prolongs animal survival. Collectively, these results demonstrate the LNP-mediated delivery of uniSTING-mRNA as a strategy to overcome the current STING therapeutic barriers, particularly for the treatment of multiple cancer types in which STING is downregulated or absent.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MicroRNA Data Integration Portal (mirDIP) was used to identify gene targets for exosomal miRNAs and can be accessed at http://ophid.utoronto.ca/mirDIP. The gene set database Hallmarks (h.all.v6.1.symbols.gmt) from the Molecular Signatures Database (MSigDB) was used in the analysis. All raw sequencing data and associated processed data files that support the findings of this study have been deposited in the Gene Expression Omnibus under accession code GSE253724 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE253724). Source data are available for Figs. 2d–g,i,j, 3b,d–q, 4b,d,f,g, 5a–g,i,j and 6b,c,e–h and Supplementary Figs. 1c, 2a, 6b–d, 8c,d, 9 and 13d–f in the associated source data file. Source data are provided with this paper.
References
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Srikanth, S. et al. The Ca2+ sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 20, 152–162 (2019).
Li, S. et al. Prolonged activation of innate immune pathways by a polyvalent STING agonist. Nat. Biomed. Eng. 5, 455–466 (2021).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Li, L. et al. Hydrolysis of 2′3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10, 1043–1048 (2014).
Shae, D. et al. Endosomolytic polymersomes increase the activity of cyclic dinucleotide STING agonists to enhance cancer immunotherapy. Nat. Nanotechnol. 14, 269–278 (2019).
Kato, K. et al. Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1. Nat. Commun. 9, 1–8 (2018).
Pan, B.-S. et al. An orally available non-nucleotide STING agonist with antitumor activity. Science 369, eaba6098 (2020).
Chin, E. N. et al. Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic. Science 369, 993–999 (2020).
Konno, H. et al. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37, 2037–2051 (2018).
Xia, T., Konno, H. & Barber, G. N. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 76, 6747–6759 (2016).
Tse, S.-W. et al. mRNA-encoded, constitutively active STINGV155M is a potent genetic adjuvant of antigen-specific CD8+ T cell response. Mol. Ther. 29, 2227–2238 (2021).
Hong, C. et al. cGAS–STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 607, 366–373 (2022).
Tu, X. et al. Interruption of post-Golgi STING trafficking activates tonic interferon signaling. Nat. Commun. 13, 6977 (2022).
Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398 (2019).
Shang, G., Zhang, C., Chen, Z. J., Bai, X.-c & Zhang, X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP–AMP. Nature 567, 389–393 (2019).
Zhao, B. et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 569, 718–722 (2019).
Wang, C., Sharma, N., Kessler, P. M. & Sen, G. C. Interferon induction by STING requires its translocation to the late endosomes. Traffic 24, 576–586 (2023).
Wang, C. et al. STING-mediated interferon induction by herpes simplex virus 1 requires the protein tyrosine kinase Syk. Mbio 12, e03228–03221 (2021).
Li, C. et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol. 23, 543–555 (2022).
Stetefeld, J. et al. Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat. Struct. Biol. 7, 772–776 (2000).
Wu, J., Dobbs, N., Yang, K. & Yan, N. Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 53, 115–126 (2020).
Barber, G. N. STING-dependent cytosolic DNA sensing pathways. Trends Immunol. 35, 88–93 (2014).
de Oliveira Mann, C. C. et al. Modular architecture of the STING C-terminal tail allows interferon and NF-κB signaling adaptation. Cell Rep. 27, 1165–1175. e1165 (2019).
Abe, T. & Barber, G. N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 (2014).
Liu, T., Zhang, L., Joo, D. & Sun, S.-C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2, 1–9 (2017).
Tak, P. P. & Firestein, G. S. NF-κB: a key role in inflammatory diseases. J. Clin. Investig. 107, 7–11 (2001).
Xu, J. et al. Precise targeting of POLR2A as a therapeutic strategy for human triple negative breast cancer. Nat. Nanotechnol. 14, 388–397 (2019).
Hotz, C. et al. Local delivery of mRNA-encoded cytokines promotes antitumor immunity and tumor eradication across multiple preclinical tumor models. Sci. Transl. Med. 13, eabc7804 (2021).
Hewitt, S. L. et al. Durable anticancer immunity from intratumoral administration of IL-23, IL-36γ, and OX40L mRNAs. Sci. Transl. Med. 11, eaat9143 (2019).
Akita, H. Development of an SS-cleavable pH-activated lipid-like material (ssPalm) as a nucleic acid delivery device. Biol. Pharm. Bull. 43, 1617–1625 (2020).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Augustine, C. et al. Blood parameters of wistar albino rats fed processed tropical sickle pod (Senna obtusifolia) leaf meal-based diets. Transl. Anim. Sci. 4, 778–782 (2020).
Marcus, A. et al. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity 49, 754–763 (2018).
Li, W. et al. cGAS-STING–mediated DNA sensing maintains CD8+ T cell stemness and promotes antitumor T cell therapy. Sci. Transl. Med. 12, eaay9013 (2020).
Krishna, S. et al. Stem-like CD8 T cells mediate response of adoptive cell immunotherapy against human cancer. Science 370, 1328–1334 (2020).
Tkach, M. & Théry, C. Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232 (2016).
Torralba, D. et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat. Commun. 9, 2658 (2018).
Ishii, H. et al. miR-130a and miR-145 reprogram Gr-1+ CD11b+ myeloid cells and inhibit tumor metastasis through improved host immunity. Nat. Commun. 9, 2611 (2018).
Yang, J. et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene 33, 3014–3023 (2014).
Ji, Y., Hocker, J. D. & Gattinoni L. in Seminars in Immunology (eds Kroemer, G. & Mantovani, A.) 45–53 (Elsevier, 2016).
Lee, S. Y. et al. Wnt/Snail signaling regulates cytochrome c oxidase and glucose metabolismregulation of mitochondria and metabolism by Wnt/Snail. Cancer Res. 72, 3607–3617 (2012).
Stemmer, V., De Craene, B., Berx, G. & Behrens, J. Snail promotes Wnt target gene expression and interacts with β-catenin. Oncogene 27, 5075–5080 (2008).
Xu, X., Zhang, M., Xu, F. & Jiang, S. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol. Cancer 19, 35 (2020).
Tokar, T. et al. mirDIP 4.1—integrative database of human microRNA target predictions. Nucleic Acids Res. 46, D360–D370 (2018).
Hashiba A. et al. The use of design of experiments with multiple responses to determine optimal formulations for in vivo hepatic mRNA delivery. J. Control. Release 327, 467–476 (2020).
Sabnis, S. et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018).
Acknowledgements
This work was supported by innovation grants (RX03202109 and RX03222104) from the Eshelman Institute for Innovation (to R.L.), a developmental grant (MCR0634222) from UCRF (to R.L.) and research grants from the NIH (R01EB032865 to R.L. and R35-CA232109 and R01-AI029564 to J.P.-Y.T.). E.M. was supported by T32-CA196589.
Author information
Authors and Affiliations
Contributions
Y.W., S.L., J.P.-Y.T. and R.L. conceived and designed the research. R.L. designed the uniSTING. Y.W. and S.L. performed plasmid construction, protein expression, mRNA synthesis, LNP generation, and all the tissue culture and in vivo animal experiments. M.H. performed RNA-seq and GSEA analysis and participated in the design of EV-related experiments. Y.Y. performed raw data alignments and analysis from RNA-seq results. E.M., L.Z. and A.M.W. helped with the preparation of mRNA and LNPs. Y.W. and M.H. performed the statistical analysis. With input from J.P.-Y.T. and S.L., Y.W. and R.L. analysed all the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Hadi Valadi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–20 and Tables 1 and 2.
Supplementary Data 1
Source Data for Supplementary Figures 1, 2, 6, 8, 9 and 13.
Source data
Source Data Fig. 1
Unprocessed western blots and/or gels.
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 6
Statistical Source Data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Li, S., Hu, M. et al. Universal STING mimic boosts antitumour immunity via preferential activation of tumour control signalling pathways. Nat. Nanotechnol. (2024). https://doi.org/10.1038/s41565-024-01624-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41565-024-01624-2
This article is cited by
-
Controlling the STING pathway to improve immunotherapy
Nature Nanotechnology (2024)