Abstract
Minimal Cut Sets (MCSs) identify sets of reactions which, when removed from a metabolic network, disable certain cellular functions. The traditional search for MCSs within genome-scale metabolic models (GSMMs) targets cellular growth, identifies reaction sets resulting in a lethal phenotype if disrupted, and retrieves a list of corresponding gene, mRNA, or enzyme targets. Using the dual link between MCSs and Elementary Flux Modes (EFMs), our logic programming-based tool aspefm was able to compute MCSs of any size from GSMMs in acceptable run times. The tool demonstrated better performance when computing large-sized MCSs than the mixed-integer linear programming methods. We applied the new MCSs methodology to a medically-relevant consortium model of two cross-feeding bacteria, Staphylococcus aureus and Pseudomonas aeruginosa. aspefm constraints were used to bias the computation of MCSs toward exchanged metabolites that could complement lethal phenotypes in individual species. We found that interspecies metabolite exchanges could play an essential role in rescuing single-species growth, for instance inosine could complement lethal reaction knock-outs in the purine synthesis, glycolysis, and pentose phosphate pathways of both bacteria. Finally, MCSs were used to derive a list of promising enzyme targets for consortium-level therapeutic applications that cannot be circumvented via interspecies metabolite exchange.
Similar content being viewed by others
Introduction
Staphylococcus aureus and Pseudomonas aeruginosa are opportunistic pathogens commonly associated with the skin microbiome and water sources, respectively. The two problematic bacteria are responsible for an estimated 1+ million deaths yearly due in part to widespread antimicrobial resistance1. S. aureus and P. aeruginosa are frequently co-isolated from chronic wounds and cystic fibrosis lungs2,3. Their interactions such as metabolite cross-feeding4,5 have been associated with higher resiliency to antibiotics and worse patient outcomes6,7. The complex nature of their interactions with other bacteria and the environment has motivated a growing number of studies involving consortia of these pathogenic bacteria, whether it is through in vivo and in vitro models8,9, or in silico models10,11. Better informed and therefore more effective intervention strategies for treating S. aureus and P. aeruginosa infections could save millions of lives and billions of dollars in healthcare expenses.
Constraint-based metabolic modelling (CBM) is a well-established systems biology field involving the computational reconstruction and analysis of biological mechanisms at multiple levels12. At its core are metabolic networks, hypergraphs described by a set of metabolites and reactions linked to each other by stoichiometric coefficients stored in a stoichiometric matrix. The constraint-based modelling approach calculates metabolite fluxes based on the assumption that the system is at steady-state; therefore, intracellular metabolite production and consumption are balanced over relevant time intervals.
Flux balance analysis (FBA) is one type of CBM that uses linear optimization to identify solutions of metabolic models, based on an objective function which often involves maximizing the flux through a biomass synthesis reaction13. The FBA solution is a flux distribution that predicts cellular phenotype including which enzymes are active and what the magnitude of the flux is for each enzyme. The biomass synthesis reaction accounts for cell growth as observed experimentally14. FBA and derived methods are used to make in silico phenotype predictions based on changes in the growth medium or based on altering of enzyme activity through gene knockouts or recombinant interventions15.
Elementary Flux Mode (EFM) analysis is another CBM method that performs an exhaustive enumeration of the edges of the metabolic solution space defined by the stoichiometric matrix; FBA solutions are nonnegative linear combinations of EFMs16. The number of EFMs grows exponentially in relation to the number of reactions; counting all EFMs has been proven to be #P-hard17. Consequently, enumeration of EFMs from large metabolic models with over 100 reactions is challenging, requiring special computational methods18, biological constraints such as transcriptional regulation19 and thermodynamic data20, and careful model network compression21,22.
Building from the set of metabolic reactions encoded in the genome, and progressing to the intricate mechanisms at the protein and enzyme level, CBM contributes to the description of a wide variety of cellular processes. Genome-scale metabolic models (GSMMs), large-scale constraint-based metabolic models computationally generated from genomes of interest are now the norm23,24, due to increased availability of data and computational power. GSMMs are well suited for identifying putative drug targets through predicting gene and metabolite essentiality25,26.
GSMMs have been applied to analyse drug targets in cancerous cells27,28 and to treat P. aeruginosa infections29. The methods identified essential reactions and synthetic lethals (SLs)30. Synthetic lethals refers to combinations of gene-deletions or enzyme interference targets which prevent growth. While the term initially referred to pairs of genes, it is now used to describe n-tuples of reaction targets. The synthetic lethals may explicitly consider both the metabolic potential of the organism and the role of the nutritional environment provided by the extracellular medium.
Improved algorithms for computing synthetic lethal strategies have been proposed to shorten the calculation process, such as Fast-SL31 and Rapid-SL32. The computation of synthetic lethals deals with a combinatorial exploration of every possible n-tuple of reactions. Thus on large networks of over a thousand reactions, computation times increase as the size of n-tuples increase, and become impracticable for n-tuples of over 4 reactions31.
Another method proposed for identifying synthetic lethals, whether n-tuple size is under or over 4, is the computation of Minimal Cut Sets (MCSs), with the biomass synthesis reaction as a target reaction. MCSs are traditionally defined as the ’Minimal Hitting Sets’ of Elementary Flux Modes (EFMs)17,33, and are an exhaustive way of exploring robustness of a network. Setting a certain reaction as target for inactivation, MCSs define all sets of reactions capable of preventing flux through the target reaction34. MCSs analysis has demonstrated remarkable performance in identifying synthetic lethals in cancer cells28. Additionally, Minimal Cut Sets have been formalized for metabolic engineering and recombinant strain optimization33.
MCSs suffer the same computational time challenges as EFMs. The number of possible MCSs grows exponentially with the number of reactions35. Interestingly, it has been proven that MCSs can be enumerated as the EFMs of a so-called dual metabolic network36. As a result, similarly to how Mixed-Integer Linear Programming (MILP) methods were developed for computing the shortest EFMs of a metabolic network37, MILP methods for computing the shortest MCSs have been developed38.
Furthermore, it might be useful to convert the obtained MCSs into sets of target genes or proteins for biological interpretation. Gene-Protein-Reaction association rules (GPRs) have been developed for GSMMs and integrated into the stoichiometric matrix39. This data has been repurposed for the MCSs computation28,40, as a way to get intervention targets.
Here, we present a new approach for calculating and analysing MCSs using our aspefm tool41. The aspefm program is a logic programming method designed to compute subsets of EFMs while respecting user-defined constraints. It differs from the Double Description method, implemented in efmtool42, which needs to enumerate all solutions before generating results, and from MILP-based methods, implemented in CNApy40,43 and CoBAMP44, which minimize the size of the reaction set. We have extended the functionality of aspefm to the computation of MCSs.
Throughout this work, we distinguish MCSs of small size (reaction n-tuples of size 3 or less including essential reactions, synthetic lethal pairs and synthetic lethal triplets) from MCSs of large size (defined here as reaction n-tuples of size 4 or more). The MCSs of small size are often the desired reaction sets, they are readily calculated by MILP methods and SLs computation algorithms, and easily converted to gene and enzyme targets using GPRs. On the other hand, the MCSs of large size are less well-studied. While they are informative of network robustness, they might be harder to compute and interpret without incorporation of biological constraints. In this study, we show aspefm was able to bridge the gap between the two types of MCSs, using large-size MCSs to reveal possible interspecies metabolite interactions.
aspefm successfully processed an aggregate model of two GSMMs totalling over three thousand reactions: a consortium model of bacteria S. aureus and P. aeruginosa. The tool efficiently identified solutions of interest in acceptable computation times despite the size of the consortium metabolic network. The aspefm application revealed potential, nonobvious, interspecies metabolite exchanges that could help the consortium growth. Promising therapeutic targets for controlling the problematic pathogens were obtained by restricting the search of enzyme targets to knock-outs that were not nullified by consortial metabolite exchanges.
Results
Overview of genome-scale metabolic models for analysis of single species and consortium
Manually curated genome-scale metabolic models (GSMM) of Staphylococcus aureus and Pseudomonas aeruginosa were selected for our analysis. The S. aureus GSMM, iYS854, was developed based on S. aureus str. JE245. The GSMM has been used for assessing the validity of experimentally determined transcriptional regulation modulons of S. aureus46. The model has been graded as the most accurate S. aureus GSMM currently available, according to a study by Renz and Dräger47.
The P. aeruginosa GSMM iPae1146 is based on P. aeruginosa strain PAO148. The GSMM was used for a high-throughput essentiality analysis29. In that study, the model was predicted to have around 97% accuracy for at predicting gene essentiality during growth on Lysogeny Broth (LB) medium.
Both metabolic models were pre-processed and curated for our analysis, as detailed in the Methods. The resulting iYS854 model includes 1454 reactions, 1338 metabolites, and 866 genes, while the resulting iPae1146 model includes 1495 reactions, 1283 metabolites, and 1148 genes.
The models were analysed in an in silico extracellular environment defined by CSP chemically defined medium49, on which S. aureus and P. aeruginosa can grow as biofilms in vitro50. The CSP medium was designed to serve as a simplified analogue of chronic wound exudate. The medium was chosen as the base for all predictions of growth and consortial cross-feeding in our study.
A consortium model consisting of P. aeruginosa and S. aureus was built from iPae1146 and iYS854 by adding metabolite exchange reactions with a shared control volume containing the growth medium. The newly created metabolic network contains 3241 reactions and 2752 metabolites. When the consortium model was constrained by an extracellular environment defined by CSP medium, a total of 57 metabolites were classified as ’external’ substrates. 46 external substrates were available to both S. aureus and P. aeruginosa, one metabolite was exclusively available to P. aeruginosa: citrate and ten metabolites were exclusively available to S. aureus including some vitamins and purines. A summary of medium metabolites is provided in Supplementary Table 2.
The network compression process, which is required for MCSs computation, excluded 1296 blocked reactions from the CSP-constrained consortium model and returned a compressed consortium network of 1062 reactions and 600 metabolites. Statistics and results from the construction and compression of the consortium model, and of the individual species models iYS854 and iPae1146, are reported in Table 1.
As well, the numbers of MCSs of small size for the consortium and individual species models are reported in Table 1. 583 MCSs of size three or less were found for the single-species iPae1146 model; 938 MCSs for the single-species iYS854 model. Two single reactions are essential to the consortium-level model, the uptake of ferrous ions and the secretion of glycolate. These two conditions are necessary for the biomass synthesis reactions of both models.
As per Fig. 1, our study relies on small-size single-species MCSs as queries to derive larger-size consortium-level MCSs. While small-size MCSs could be computed using existing MCSs tools or the Synthetic Lethals algorithms, there are differences in computational performance between aspefm and the other MCSs computation tools as summarized in Supplementary Table 1. These differences motivate the continued study of MCS algorithms to identify best practices for specific scientific inquiries.
MCSs of size three or less are denoted as small-size MCSs. a Performance of MCSs computation tools on the S. aureus and P. aeruginosa consortium model is tested with consortium-lethal MCSs. b Small-size MCSs are computed on single-species models, then tested for growth recovery on the consortium-level model. c For MCSs which are no longer lethal on the consortium model, larger size consortium MCSs are computed, revealing metabolite exchanges. d For MCSs that are still lethal on the consortium retrieved in both bacteria, interspecies protein structure alignments of AlphaFold structures are performed, and the best drug targets for therapeutic intervention in human are retrieved.
Single-species MCSs are classified based on their ability to rescue growth in the consortium model. In a prelimlinary step, performance of aspefm was compared to MCSs tools CNApy43 and CoBAMP44 for computation of large-size solutions on the consortium model. In a second step, single-species MCSs that rescued growth were used to derive the exact metabolite exchanges that nullified the cut set from the consortium model through a constrained consortium MCSs analysis. As a final step, single-species MCSs that did not rescue growth were used to develop a ranking of consortium-level enzyme targets based on protein structure similarity.
For the final step, single-species MCSs that did not rescue growth and were identical in both microorganisms were collected and gathered into larger reaction sets. These reaction sets are, by definition, consortium-level cut sets. This approach was deemed better than a constrained MCSs analysis on the consortium model for two reasons. First, MCSs tools could not perform unconstrained enumeration of small-size MCSs of the consortium model in reasonable time and second, MCSs at the consortium-level mostly consisted of combinations of single-species MCSs. Enzyme targets were derived from cut sets that could correspond to single enzyme pairs.
aspefm efficiently calculates MCSs from consortium-level models regardless of the size of the reaction set
The performance of our aspefm tool was evaluated by computing MCSs from the compressed consortium model comprised of 1062 reactions. The target reactions for the simulation were the biomass synthesis reactions for both single species. The only constraints for the simulation were to identify MCSs of reaction size 16 or less and a maximum run time of 1.5 days.
The performance of aspefm was compared with CNApy43 and CoBAMP44, both are MILP-based MCSs enumeration methods. For each tool, five executions were launched and averaged (Fig. 2). On average, aspefm identified more than thrice as many compressed MCSs as CNApy in the 1.5 day run time. aspefm averaged 3663.8 MCSs while CNApy averaged 1213.0 MCSs. Both aspefm and CNApy were able to enumerate MCSs regardless of the reaction set size, while CoBAMP was heavily hindered by its forced iterative enumeration approach which started with the smallest cut sets; the method only identified around 203.6 MCSs on average. Upon decompression, the MCSs found by aspefm and CNApy reached the order of 105 MCSs, illustrating the necessity of network compression.
The MCSs identified using aspefm sampled solutions ranging from 1 to 16 reactions with the highest frequency at a reaction size of 5 (Fig. 2). As illustrated by the high variability over 5 executions, no substantial preference was observed towards any particular reaction size: aspefm is a logic programming tool that benefits from the combinatorial exploration nature of its SAT-based capabilities. Meanwhile, the MCSs identified using CNApy were biased toward smaller reaction sizes and mainly enumerated solutions with 2–7 reactions, with the highest frequency occurring at 3 reactions (Fig. 2). These results from the consortium model suggest that in contrast to CNApy and especially CoBAMP, aspefm enumerated solutions independently of their size. For computing MCSs of large size from the consortium model, aspefm is thus the tool of choice.
Consortium-level MCSs reveal role of interspecies metabolite exchanges in single-species growth
Medical infections comprised of both S. aureus and P. aeruginosa can result in worse patient outcomes and can be more difficult to treat than monocultures. Thus therapeutic treatments ideally should not only target a single species, but the whole consortium. Metabolic modelling can identify metabolite exchanges between species that would bypass therapeutic strategies targeting only a single species. MCSs of small size from the single-species models were tested for lethality at the consortium level to determine if directed cross-feeding interactions or passive metabolite exchange through metabolite leaking could circumvent single-species lethalities. Of the 583 MCSs of size three or less for P. aeruginosa, 68 were no longer cut sets at the consortium level based on metabolites secreted by S. aureus. Meanwhile, of the 938 MCSs of small size for S. aureus, 199 cut sets were no longer effective due to metabolite exchanges from P. aeruginosa.
The lethal MCSs that were nullified due to metabolite exchanges were further analysed with the consortium-level model. aspefm was chosen for determining the identity of the exchanged metabolites, since the necessary computation involved consortium-level MCSs of large size: over three reactions for the most part. Constraints for the computation were separated into two categories. We defined within aspefm: ‘wanted reactions’ (positive Boolean inputs) for reactions that should appear in the MCSs, and ‘unwanted reactions’ (negative Boolean inputs) for reactions that should not appear in the MCSs; the latter should not be confused with the target reaction. aspefm was run on the consortium model for each nullified MCS and for each bacterium, the reactions from the invalidated MCS were set as the ‘wanted reactions` and all reactions unnecessary for metabolite exchange as the ‘unwanted reactions’; the target reaction was set to the single-species biomass reaction (see Methods). As a result, generated consortium-level MCSs were composed only of the previously computed single-species MCSs and of exchange reactions, revealing the metabolites essential for recovering growth.
The obtained MCSs were limited to eight or fewer reactions and a time limit was set to 1.5 days for each computation. In total, 531 compressed consortium model MCSs were computed, ranging in size from 2 to 8 reactions, with the mean and median being 6 reactions and the highest frequency being 7 reactions. The details of each consortium-level MCS identified through this analysis are provided in Supplementary Dataset 1.
The identity of the exchanged metabolites in the consortium model and the number of single-species MCSs they suppress are reported for each bacterium in Fig. 3. The same single-species MCSs could be invalidated by the exchange of any one of several metabolites. Indeed, for a given single-species MCS, several consortium MCSs could be found, indicative of intervention strategies at the consortium level, and corresponding each to one or several metabolites.
For instance, a MCS of size three that exists for a single-species model might be nullified by any of five different metabolite exchange reactions, together in a consortium model MCS of size eight. Consequently, at least five interventions on exchanges with the other bacterium would be required for the original MCS to regain lethality. Alternatively, if among all consortium MCSs, there was only one metabolite exchanged to counter the single-species MCS, then only a single theoretical intervention would be required.
The majority of cut sets nullified due to metabolite exchanges involved purine metabolism, pentose phosphate pathway, and glycolysis. Inosine was a pivotal metabolite in many of those functions, it was able to complement almost half of the identified cut sets for each bacterium. Inosine nucleosidase can transform the metabolite into hypoxanthine—a purine – and ribose, which can support many central metabolism transformations.
Purines xanthine and hypoxanthine seemed to play a central role in metabolite exchanges allowing the recovery of growth, likely due to their relationship with nucleotide metabolism. Other notable metabolite exchanges shared by the two bacteria include acetylglucosamine, ribose, fructose and glycerol, complementing glycolysis functions; and urea-related metabolites, complementing urea cycle metabolism functions.
Previously, the enzyme N5,N10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase, catalysing methylenetetrahydrofolate dehydrogenase and methenyltetrahydrofolate cyclohydrolase, has been studied as potential drug target for P. aeruginosa51. However, these reactions, identified as essential on the single-species model, are complemented by purines and histidine exchanged by S. aureus on the consortium-level model.
Although the majority of the single-species level MCSs corresponded to large-size consortium-level MCSs: most recovering growth by at least two distinct possible metabolite exchanges and making for very impractical treatments, we found a few small-sized MCSs, such as glucosamine-6-phosphate synthase with acetylglucosamine exchange.
Interestingly, while glucosamine-6-phosphate synthase was identified as an essential reaction in both single-species models, the lethal phenotype conferred by deleting this reaction would be recovered by a simple exchange of acetylglucosamine between the bacteria. Therefore, efforts using therapeutic agents which inhibit this enzyme52 would need to consider the potential role of acetylglucosamine found in the environment or leaked by resistant bacteria in the consortia.
Consortium-level MCSs identify multi-species, structurally close, intervention targets
The effects of therapeutic agents that target a single species can, in some cases, be bypassed through metabolite exchange from other consortia species. From single-species MCSs, our analysis illustrated how specific consortium-level metabolite leakage or cross-feeding events could enable growth recovery. This identified MCSs that should be excluded from further analysis, while the remaining MCSs could be more promising targets for therapeutic intervention.
In order to retrieve consortium-level intervention targets, we selected MCSs that were mutually shared by the two bacteria. As well, the corresponding enzyme targets were retrieved from MCSs for protein structure studies. It is hypothesized that proteins with similar structure could be targeted simultaneously by the same therapeutic agent and thus target the consortium by conjointly blocking both S. aureus and P. aeruginosa. Out of 515 iPae1146 MCSs and 739 iYS854 MCSs of small size that were not nullified by metabolite exchanges, 65 MCSs were shared by both bacteria (Table 1).
The structures of the proteins associated with the shared 65 MCSs were analysed for similarity. The protein structure analyses ruled out several MCSs since the target enzymes for S. aureus and P. aeruginosa were not deemed similar enough for concurrent therapeutic intervention. Enzymes were represented by their GSMMs gene assignments, ie. GPRs (Gene-Protein-Reaction gene products), and were retrieved from MCSs with a logic programming extension of our aspefm procedure. Interspecies relative enzyme structure similarity was measured by Root Mean Square Deviation (RMSD) of atom positions in Ångström, protein structure alignments were computed on AlphaFold structure predictions53. The remaining shared MCSs were filtered using an additional criterion: the S. aureus and P. aeruginosa protein structures were compared to any potential human homologues. MCSs with enzymes that were deemed too similar to human homologues were removed. As a result, a ranking of the most promising MCSs for antibiotics discovery in human is presented in Fig. 4.
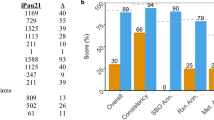
Only P. aeruginosa and S. aureus targets whose protein structures matched best were kept. a Ordinates labels show targetable enzymes. Bars heights are Root Mean Square Deviation (RMSD) of atom positions, measured in Ångström, resulting from protein structural alignment of enzyme pairs. Enzymes indicated with ‘H’ were found to have human homologues. b Pie distribution for associated metabolism groups for each targetable enzyme in a. c Pie distribution of categories of MCSs potentially targetable in human. From the 23 MCSs targetable in human (in c), only 12 are independent of the growth medium (shown in a and b).
From the 65 MCSs common to both bacteria and their corresponding enzymes, 23 of them were considered good potential drug targets for elimination of the bacterial consortium in human (Fig. 4c). Details and ranking of each MCS are provided in the Supplementary Dataset 2. The most promising enzymes to simultaneously target both S. aureus and P. aeruginosa included twelve enzymes from nucleotide metabolism, lipid synthesis, aromatic amino acid biosynthesis, bacterial cell wall construction, amino sugar metabolism and folate biosynthesis (Fig. 4a, b).
At the top of the ranking, beta-ketoacyl-ACP synthase III had high protein structure similarity between the two bacteria (interspecies RMSD: 1.21 Å) and fortuitously, there did not seem to be human homologues. However, there exist functionally related isozymes beta-ketoacyl-ACP synthase I and II, presenting high similarity with human homologues, which were thus excluded by our procedure.
Some example cell wall synthesis enzymes include undecaprenyl-disphosphatase (interspecies RMSD: 1.37) and UDP-N-acetylmuramoyl-L-alanine synthetase (RMSD: 1.81). Both enzymes are found only in the bacteria and not in human. Amino sugar metabolism included two enzymes for which there are human homologues: GlmU and phosphoglucomutase, however the protein structures were considered dissimilar enough by our filtering procedure for these enzymes to be considered targetable.
Other enzymes with high therapeutic potential to treat S. aureus and P. aeruginosa consortia are four enzymes from the aromatic amino acid biosynthesis pathway: chorismate synthase (RMSD: 1.22), 3-phosphoshikimate 1-carboxyvinyltransferase (RMSD: 1.35), shikimate kinase (RMSD: 1.91) and 3-dehydroquinate synthase (RMSD: 2.03). This biosynthesis pathway is famously not present in mammals. In fact, an inhibitor of 3-phosphoshikimate 1-carboxyvinyltransferase, glyphosate, is commonly used as an herbicide54.
Additionally, eleven environment-dependent, bacterial-only enzyme targets were identified (Fig. 4c). These enzymes are derived from 2–3 reaction MCSs containing a transporter for an amino acid found in the growth medium and an amino acid biosynthesis reaction, which becomes essential in absence of that amino acid. Were the bacteria to be grown in an environment lacking the amino acids associated with these MCSs, an inhibitor of the enzyme targets would be effective.
These MCSs provide detailed, systemic insight into which amino acid biosynthesis reactions are the most important for S. aureus and P. aeruginosa growth, and therefore which amino acids biosynthesis pathways are the most promising targets. Our eleven MCSs correspond to six important amino acids, among two classes, aromatic amino acids (tryptophan, histidine, phenylalanine) and branched-chain amino acids (isoleucine, leucine, valine). These essential pathways are in accordance with previous studies and might be druggable55. More insight into the growth medium dependent MCSs, as well as structure alignments and human homologues, is given in Supplementary Material, Supplementary Figs. 2 and 3.
Discussion
aspefm is a powerful and promising tool for metabolic systems analysis, able to compute EFMs while respecting any applied constraints, whether they are logical or linear, and it can be extended at will by the clingo Python interface56. Here, we extend the tool to the computation of MCSs from metabolic models of single and multispecies systems. MCSs are mathematical objects with great engineering, ecological, and therapeutic potential as they identify combinations of reactions within large, highly connected networks that have outsized abilities to influence phenotype. As illustrated in Fig. 2, exploration of MCS solutions in genome-scale models is now possible. Compared to other methods, aspefm showed substantially better performance for enumeration of MCSs of large size on the consortium model within a time frame of 1.5 days. Note that the enumeration of these subsets of solutions with aspefm is non-ordered and non-deterministic, as demonstrated by the variability in MCSs sizes. In contrast, CoBAMP’s enumeration algorithm is ordered, consisting of computing the next smallest MCSs at each iteration. CNApy proposes both ordered and non-ordered enumeration, aspefm was compared to its non-ordered enumeration. With its innovative SAT-based solving, our tool aspefm was able to enumerate solutions on a consortium-level with ease, despite the network comprising about three thousand reactions.
In order to make full use of the enumeration capacities of our aspefm tool, we devised a MCSs analysis to study potential cross-feeding interactions within the consortium. Our analysis proposed the usage of MCSs of small size on single-species models as constraints for the computation of MCSs of large size on a consortium-level (Fig. 1). Single-species MCSs were tested for the recovery of growth on the consortium model, and if metabolite exchanges permitted growth recovery, then MCSs of large size were computed, providing insight into the metabolite exchanges in question (Fig. 3). To do so, all reactions that were not metabolite exchange reactions were set as negative Boolean inputs, and MCSs of small size were used as positive Boolean inputs, and aspefm was run with a time limit of 1.5 days. We implemented an additional FBA check for all MCSs identified by the solver to verify the minimality of the solutions. Therefore we present the utility of an exhaustive yet constrained metabolic pathways analysis, through application of biological relevant constraints, as we have presented previously for EFM analysis41. Rather than a complete enumeration of all cut sets, which like EFM enumeration is only achievable on modestly sized reaction networks, we enumerate a subset of MCSs while answering a specific biological question, under a finite time limit.
This new application of MCSs highlights the potential role of metabolite exchanges and metabolite cross-feeding on consortium functioning and resilience to therapeutic efforts (Fig. 3). We strengthened the study by combining the analysis with drug target predictions for the single-species MCSs that did not regain growth with consortial metabolite exchanges (Fig. 4). Meylan and coauthors have showed strong evidence that antibiotic tolerance might be affected by the impact of metabolite exchanges, in particular they showed that uptake of fumarate/glyoxylate by P. aeruginosa increases/decreases respectively its tolerance to aminoglycosides57,58. Usually, MCSs of size three or fewer are considered the most biological relevant. This logic follows the argument that disrupting more than three different genes, or interfering with more than three mRNA targets or the drugging of more than three enzymes at the same time is challenging. MCSs of small size are readily enumerated from single-species metabolic models, and can be performed with existing SLs enumeration tools or with MCS enumeration tools. However, we argue that MCSs of large size can provide valuable information on network robustness and fragility. MCSs of large size cannot be reliably enumerated by SLs tools as these are iterative methods not adapted for combinatorial enumeration. Here, MCSs of large size revealed the explicit metabolite exchanges between the two bacteria that enabled recovery of growth after three or fewer reactions were cut. We believe this methodology can be extended to larger aggregate models including microbiome-level systems and be used to systematically identify metabolite exchanges that can and can not circumvent medical treatments.
We validated the our predicted metabolite exchanges uncovered with MCSs using COBRAPy and FBA to test all possible metabolite cross-feeding interactions; the results are reported in Supplementary Table 3. Of note, metabolite exchanges can also be retrieved on bacterial consortia models through using EFMs analysis59,60. Interestingly, we compared our tool to SMETANA, a tool for estimating growth-dependent species exchanges in bacterial consortia61. 25% of our highlighted metabolite exchanges were not predicted by SMETANA. The analysis performed by SMETANA on our consortium model is given in Supplementary Fig. 4. We believe interspecies metabolite exchanges are representative of at least three possible mechanisms: cross-feeding (bacterial co-operation), metabolite leakage (non specific loss of metabolites necessary for biomass production), and metabolite acquisition from the growth medium which may include necromass62.
Our study positions itself in recent efforts from the metabolic modelling community in bringing metabolic models to the cellular consortium level63. There is a growing interest in multi-species models such as AGORA for modelling of gut microbiota64, and multicellular models such as whole-body human models65. However, we found that even the well-curated single-species models presented in our study were not exempt from curation issues. For example, iPae1146 lacked reactions for aminoacyltransferase reactions. Interestingly, the aminoacyltransferases reactions were present on the smaller P. aeruginosa PAO1 model iMO1056 from 200866. Another curation issue included the lack of a functioning formate transporter for iPae1146 and of a citrate transporter for iYS854, even though these metabolites might have substantial interspecies exchange potential.
Data from UniProt and AlphaFold53 was used to predict therapeutic targets based on protein structure similarities. Protein structure data was not available for many of the enzymes of interest which limited the mapping of the specific protein sequence to a model structure. We therefore used the application to quantify similarities of the predicted structure of two protein sequences rather than the overall accuracy of the 3D structure. If the predicted protein structures were deemed similar enough, it is hypothesized that an inhibitor would be more likely to be a ligand to both. AlphaFold derived preemptive ranking for further analysis and provided a uniform method of applying the computational workflow. Enhanced predictions will be possible with improved crystallization-based protein structures, and through further analysis of the enzyme targets, their active sites, and medically approved inhibitor ligands67.
Finally, a relevant point of discussion is the conversion of enzymes to gene and reaction data, ie. Gene-Protein-Reaction rules (GPRs). GPRs are composed of AND and OR Boolean rules, symbolizing complexes and isozymes respectively; GPRs are associated to the reactions they catalyse. Previous studies have incorporated GPRs into the stoichiometric matrix39,40. This has the downside of forcing flux through artificial reaction constructs. In contrast, our approach for converting reactions to proteins using GPRs made use of logic programming, – as GPRs are Boolean logic rules – thus further expanding the aspefm framework. GPRs are of major importance when analysing the enzymes gene or protein data, and were useful for us when retrieving AlphaFold entries53.
To conclude this study, we argue that biomass-associated MCSs of small size and of large size were equally useful. With our method, a subset of all large sized MCSs was able to reveal consortium-level metabolite exchanges that could only be observed after analysis of small sized MCSs. Additionally, MCSs which could not be complemented based on interspecies metabolite exchanges were analysed for their role as drug targets. We thus propose that there is a strong need for MCSs enumeration tools such as aspefm, and for metabolic modelling methods as a whole in the context of microbial ecology, medical intervention, and drug discovery.
Methods
Minimal Cut Sets were computed on Staphylococcus aureus and Pseudomonas aeruginosa GSMMs iYS854 and iPae1146, as well as on a consortium model containing the two models and reactions to model cross-feeding. The tools used are aspefm, our tool, for which we detail the methods further, CNApy and CoBAMP. For representing CNApy, the StrainDesign Python module was chosen and configured with the non-ordered enumeration algorithm68. We provided code for the analysis at https://github.com/maxm4/paSAmcs/. The repository includes the models and scripts for running every tool. We used IBM © cplex for linear programming solving and an Intel® Xeon® E5-2609v2 2.5GHz processor.
An overview of the complete analyses performed in Table 1, Figs. 3 and 4 is found in Fig. 1. MCSs of small size of P. aeruginosa and S. aureus were tested for growth recovery on the consortium model. For MCSs which lost their lethal phenotype on the consortium, MCSs of large size were retrieved explaining which metabolite exchanges allow recovery of growth. For the remaining MCSs of small size, MCSs in common between the bacteria were retrieved and analysed for the search of possible new antibacterial agents. This analysis is further illustrated in Supplementary Material: Supplementary Figs. 1, 5, and 6.
Metabolic models pre-processing and curating
Both models went into a first phase of pre-processing. Erroneous identifiers were resolved and renamed. A pyocyanin transporter was added to iPae1146, as P. aeruginosa is known to secrete pyocyanin in presence of S. aureus10. As well, new transporter reactions were added to iYS854 to match the Chemically Defined Medium used in ref. 69, and such that the well-studied WTA-null non-lethal Staphylococcus aureus mutant Δ tarO could be accounted for, in order to resolve the ambiguities raised by ref. 45.
Accuracy of the models was estimated using the MEMOTE community tool for assessing GSMM quality70. iPae1146 scored low at 23 %, mainly due to its lack of annotations, while iYS854 scored at 75%. In addition, network topology issues were reported in Table 1. The iPae1146 model contains a smaller number of exchange reactions, about twice as many blocked reactions, and a substantially higher number of reactions implicated in stoichiometrically balanced cycles.
As well, 29 extracellular metabolites from iPae1146 and 38 from iYS854 were found to lack exchange reactions. Notable metabolites from these lists, which are thus excluded from the possible metabolic interactions between bacteria, include formate for iPae1146 and citrate for iYS854.
The models were constrained to CSP Chemically Defined Medium50. For all 47 CSP medium metabolites metabolized by P. aeruginosa and all 56 CSP medium metabolites metabolized by S. aureus, exchanges lower flux bounds were set to flux values in accordance with their relative quantity in the medium. The obtained constrained metabolic models are presented in Supplementary Dataset 3, in the standard SBML format.
Consortium model construction and analysis
A consortium model of P. aeruginosa and S. aureus models was constructed using exchange reactions of both models as means for cross-feeding. All reactions, metabolites and compartments of the original models were subtitled with ‘PA’ or ‘SA’ in the consortium model depending their origin. The consortium model’s highest level compartment permited exchange of metabolites between P. aeruginosa and S. aureus, as illustrated in Fig. 5.
Additional boundary exchange reactions were added to all newly created extracellular metabolites using COBRAPy71, and the new exchange reactions fluxes were constrained to correspond to metabolites of the CSP Chemically Defined Medium50. All reactions that were previously exchange reactions of the iPae1146 and iYS854 GSMMs have become reactions that can be used for cross-feeding. We set arbitrary flux bounds of [−20, 20] for all cross-feeding reactions, in accordance with the minimum possible uptake flux in CSP Medium, which is set to −20 mmol/gDW/hr for O2, as is standard for that metabolite in COBRA models. The resulting consortium metabolic model in SBML is presented in Supplementary Dataset 3.
Supplementary Table 3 presents the possible metabolite exchanges between the bacteria, as identified with COBRAPy tests. Within the consortium model, P. aeruginosa can metabolize 36 metabolites of the 73 metabolites potentially secreted by S. aureus, while S. aureus can metabolize 47 metabolites of 65 metabolites potentially secreted by P. aeruginosa. These metabolites are reported in Table 1 as well.
Note that for computation of MCSs—and thus essential reactions, synthetic lethal pairs and triplets—on a model alone, its biomass is taken as the target reaction. As such, when looking for which cross-feeding reactions complement a cut set of P. aeruginosa or S. aureus alone (Fig. 3), only one of the two consortium biomass reactions should be taken, the targeted biomass reaction in question. The procedure for computing MCSs revealing cross-feeding interactions with aspefm, from the sets of lethal MCSs with growth recovery of either bacterium, is illustrated in Supplementary Fig. 6.
Separately, for modelling growth on the consortium model, the FBA objective reaction is defined as the sum of both biomass reactions. Thus, for computation of MCSs which are lethal to the whole consortium model (Fig. 2), both biomasses were taken as the target reactions.
Network compression
To help with computation efficiency, models are compressed using the network compression procedure developed by the von Kamp team, as part of the pip package efmtool_link from the Klamt lab (https://github.com/cnapy-org/efmtool_link). The tool relies on excluding blocked reactions and correcting reversibilities through Flux Variability Analysis13, then applying a nullspace-based compression method from EFMTool42.
The general principle behind nullspace-based compression was introduced in METATOOL21, and later re-explored in ref. 22. Linearly dependent lines from the stoichiometric kernel are regrouped into a single (lumped) reaction. This allows reactions that always operate together, i.e., their fluxes are linearly dependent to each other, to be regrouped into the same reaction subsets. For MCSs analysis, the linear coefficient factor between reactions in the same subset is of no importance, thus MCSs decompression is trivial.
Network dualization
As described in Ballerstein et al.36, it is possible to describe the problem of computing MCSs as the problem of computing particular EFMs on a dual metabolic network, meaning that the original network has to undergo a dualization conversion procedure.
We formulated the problem by making use of the MILP version proposed by von Kamp and Klamt in 201438, which excludes some of the linear variables and constraints introduced by Ballerstein. A notable feature of this method was defining an inequality constraint instead of an equality constraint for metabolites of the dual network that were originally irreversible reactions.
All reversible dual reactions are split into two irreversible dual reactions. As in the formalisms defined by Ballerstein and von Kamp, the reactions corresponding to reversibility constraints are the only ones to which subset-minimality applies, meaning the other linear variables are free to be either strictly positive or equal to zero following whether it suits the linear program. We provide Supplementary Fig. 7 to explain network dualization with aspefm.
Dual metabolic network formalization
Let us define S the stoichiometry matrix of size m × r, m being the number of metabolites in the metabolites set \({{{\mathcal{M}}}}\) and r being the number of reactions in the reactions set \({{{\mathcal{R}}}}eac\). Let us define the set of reversible reactions \({{{\mathcal{R}}}}ev\subset {{{\mathcal{R}}}}eac\) and the set of target reactions \(t\subset {{{\mathcal{R}}}}eac\) to be disabled for MCSs computation.
In the dual metabolic network, original reactions become metabolites, and original constraints become reactions. If S is the primal stoichiometry matrix is of size m × r, with I the identity matrix of size r × r and −T vector of size r × 1 with values 0 for j ∉ t and − 1 for j ∈ t, then \({{{\mathcal{D}}}}\) is a dual matrix of size r × d, with d = m + r + 1, and defined as \({{{\mathcal{D}}}}=({S}^{T}{{{\bf{I}}}}-{{{\bf{T}}}})\).
The computation of the dual network \({{{\mathcal{D}}}}\) generates a metabolites set \({{{{\mathcal{M}}}}}_{dual}\) and a reactions set \({{{\mathcal{R}}}}ea{c}_{dual}\). The set \({{{{\mathcal{M}}}}}_{dual}\) is simply the reactions set \({{{\mathcal{R}}}}eac\) and is thus of size r, while the reactions set \({{{\mathcal{R}}}}ea{c}_{dual}\) is of size d and composed of three different types of reactions: \({{{\mathcal{R}}}}ea{c}_{dual}={{{\mathcal{S}}}}\cup {{{\mathcal{RC}}}}\cup {{{\mathcal{T}}}}\).
More precisely, all m stoichiometry constraints \({{{\mathcal{S}}}}\) become reversible reactions, r reversibility constraints \({{{\mathcal{RC}}}}\) become reversible if and only if the original reaction is reversible too, while the reactions t to be disabled become one irreversible target reaction \({{{\mathcal{T}}}}\). More precisely, all m stoichiometry constraints \({{{\mathcal{S}}}}\) become reversible reactions, r reversibility constraints \({{{\mathcal{RC}}}}\) become reversible if and only if the original reaction is reversible too, while the reactions t to be disabled become one irreversible target reaction \({{{\mathcal{T}}}}\).
After splitting the d dual network reactions into k irreversible reactions, the set \({{{{\mathcal{R}}}}}_{dual}\) of irreversible reactions is obtained. For clarity, k = 2m + r + ∣Rev∣ + 1. For MCSs computation, we are concerned with its subset of interest \({{{\mathcal{C}}}}ut\subset {{{{\mathcal{R}}}}}_{dual}\), corresponding to the split of reactions \({{{\mathcal{RC}}}}\). The set \(Re{v}_{dual}:{{{{\mathcal{R}}}}}_{dual}\times {{{{\mathcal{R}}}}}_{dual}\to {{{\mathcal{R}}}}ea{c}_{dual}\) keeps track of which reactions of \({{{{\mathcal{R}}}}}_{dual}\) were originally reversible in the reaction set \({{{\mathcal{R}}}}ea{c}_{dual}\). This is the set of forwards backwards reaction pairs. Further ahead, we denote by D the dual matrix of size k × r after splitting reversible reactions.
Minimal Cut Sets formalization
Given \({{{\mathcal{R}}}}ev\subset {{{\mathcal{R}}}}eac\) the set of reversible reactions of the primal network, D the dual matrix of the stoichiometric matrix S, \({{{{\mathcal{M}}}}}_{dual}\) the set of dual metabolites, originally reactions of the primal network, \({{{{\mathcal{R}}}}}_{dual}\) the set of k irreversible dual reactions, \(Re{v}_{dual}\subset {{{{\mathcal{R}}}}}_{dual}\times {{{{\mathcal{R}}}}}_{dual}\) indicating which pairs of reactions result from a split, \({{{\mathcal{C}}}}ut\subset {{{{\mathcal{R}}}}}_{dual}\) the subset of reactions corresponding to directionality constraints of the primal network, and \({{{\mathcal{T}}}}\in {{{{\mathcal{R}}}}}_{dual}\) a target reaction associated to one or several primal network reactions t that should be disabled, the Minimal Cut Sets problem can be defined as the following:
Problem: Find non trivial subset-minimal assignments to {True} of \({c}_{\overline{r}}\in {\mathbb{B}}\), \(\forall \overline{r}\in {{{\mathcal{C}}}}ut\) such that :
\({D}_{\overline{m}\overline{r}}\) denotes the dual matrix D stoichiometry coefficient associated to dual metabolite \(\overline{m}\in {{{{\mathcal{M}}}}}_{dual}\) and dual reaction \(\overline{r}\in {{{{\mathcal{R}}}}}_{dual}\). Equations (1) and (2) represent the steady-state constraint, and is an equality or an inequality whether the metabolite the constraint applies on was originally a reversible or an irreversible reaction. Equation (3) is defining that all reaction fluxes \({v}_{\overline{r}}\) should be positive or null. Equation (4) associates Boolean indicator variables \({z}_{\overline{r}}\) to active reaction fluxes, meaning reaction with non-null fluxes. Equation (5) defines specific Boolean indicator variables \({c}_{\overline{r}}\) for reaction fluxes corresponding to reactions in \({{{\mathcal{C}}}}ut\). These are the Boolean variables that are considered for subset-minimal solutions. The \({{{\mathcal{C}}}}ut\) reactions are the only reactions which flux is of interest: representing the actual reactions in MCSs. Equation (6) forbids the flux of two irreversible reactions issued from the split of a reversible one to be non-null. Equation (7) defines the domain of reaction fluxes \({v}_{\overline{r}}\forall \overline{r}\in {{{{\mathcal{R}}}}}_{dual}\) as real linear values, the domain of indicator variables \({z}_{\overline{r}}\forall \overline{r}\in {{{{\mathcal{R}}}}}_{dual}\) as Boolean logic values, and forces the target reaction flux to be non-null.
Taking all of these constraints and searching for subset minimal assignments of \({c}_{\overline{r}}\) to {True}, we obtain the MCSs disabling reaction targets \({{{\mathcal{T}}}}\). Information for dual metabolic network construction and the Minimal Cut Sets problem is summarized visually with a complete formalization for genome-scale metabolic models in Supplementary Fig. 7.
Using aspefm to compute MCSs
The procedure is applied using aspefm, our Answer Set Programming (ASP) logic programming method41. The aspefm tool is distributed at https://github.com/maxm4/aspefm.
Given the compressed metabolic network in dual form, aspefm defines a logic program in Answer Set Programming able to enumerate all or only a subset of Minimal Cut Sets. ASP is a logic programming language, meaning it requires a declarative logic program to be defined as input. It is adapted for finding solutions to combinatorial problems thanks to its SAT-based solving. SAT refers to the well-studied Boolean satisfiability problem. The solver utilized in aspefm is state-of-the-art solver clingo extended with linear constraints through a modified clingo[LP] interface, with an IBM © cplex backend72.
Subset-minimal solutions are obtained with the set minimization heuristics of clingo, aspefm’s solver. No minimization of the size of the solutions searched is performed. An algorithmic amelioration was made to the clingo[LP] code. The algorithm for finding core conflicts from conflicting linear constraints was formerly implemented in recursive pure Python. We replaced it by cplex’s internal conflict refiner function. The code runs about 10-times faster on small and on larger models.
Another amelioration was made for direct enumeration of solutions with linear constraints, which are known to modify the solution space and thus its minimal solutions73. A solution checker was implemented, and it is called to verify the minimality and validity of solutions, in the case of MCSs with biomass as a target, it is a simple FBA call. This linear programming call should be very fast in computation time compared to the overall cost of the combinatorial exploration.
As with EFMs computation, additional constraints can be added to aspefm, only yielding a subset of all possible solutions. These constraints need to be expressed on the reactions from the compressed dual network, or undergo a conversion process if these relate to the original network.
Answer set programming
Answer Set Programming (ASP) is a particular specification of logic programming. It is a widely used method for combinatorial problems: it has been applied to solve various biological problems, including problems related to reconstruction of constraint-based modelling networks74,75. The Answer Set Programming logic programming paradigm is oriented towards resolving constraint satisfaction problems, combinatorial optimization applications, and NP-hard problems in general. As with other logic programming methods, it defines declarative, automated reasoning programs in human-readable syntax, for which resolution is left to the computing machine. In ASP’s case, a set of solutions can be derived, and solutions are called answer sets. The language defines the so-called stable models semantics, where a model is a solution if and only if it is stable, and thus, answer sets are also called stable models. Answer sets are analogous to truth assignments of Boolean propositions, in regards to testing the satisfiability of a Boolean formula76. We recommend Lierler’s review as both an entry-level paper for experimented SAT modellers and a comprehensive look at the Answer Set Programming field76.
Adding constraints to aspefm
Using aspefm’s input format, it is possible to add constraints to the computation of MCSs. Let us consider \({{{\mathcal{C}}}}ut\) the set of all reactions, and cr Boolean variables representing if a reaction is cut or not. For example, we can express a size constraint, meaning that cut sets above a certain size P will not be computed:
Supposing we have a non-empty list of “unwanted reactions” of interest \(U\subset {{{\mathcal{C}}}}ut\). The following negative Boolean constraint can be added:
This will specify to the solver to only compute MCSs containing none of those reactions.
Supposing we have a non-empty list of “wanted reactions” of interest \(W\subset {{{\mathcal{C}}}}ut\). The following positive Boolean constraint can be added:
This will specify to the solver to only compute MCSs containing all of those reactions. Note that since these positive Boolean inputs translate into adding linear constraints changing the solution space, our solution checker implemented in aspefm should be called.
Retrieving information at the gene and protein level
Gene and protein level information was reviewed as part of the model curation process. Gene products defined in the SBML models were modified to all match UniProt or TrEMBL entries, in particular for iYS854. iYS854 genes used new locus tags that yielded no UniProt query results, so genes were renamed to their old locus tags. UniProt entries were sought for using UniProt BLAST. For the gene products with no match in the str. JE2 strain, an homologue from a close S. aureus strain was used. In the process, new RDF annotations were added for future modellers. For both models, overall Boolean Gene-Protein-Reaction rules (GPRs) were simplified into canonical Disjunctive Normal Form (DNF), which helps with exhaustive enumeration of gene knock-outs from sets of reactions. Minimal sets of genes corresponding to the MCSs were computed using ASP logic programming with subset-minimization heuristics, as the problem can be expressed as minimal Boolean assignments to {True}.
Minimal sets of genes are non-trivial subset-minimal assignments to {True} respecting the GPRs Boolean formulae. However, as underlined in Machado et al.39, some gene products are ubiquitous, appearing in multiple reactions, meaning that the minimal sets of genes obtained from lethal MCSs of reactions might not necessarily be minimal in the number of genes to be knockout.
In order to better describe information at the gene or protein level, it is required that each reaction is associated to at least one gene. So, for transporter reactions that are not associated to genes, we added a dummy association, which in fact corresponds to the either backwards or forwards direction of that reaction, since reactions are split for MCSs computation. In practice, almost all reactions lacking genes are transporters, and forwards transporter reactions should also be assumed to be dependent on the presence of external metabolites in the medium. Some transporter reactions are also known to be spontaneous and annotated as such in their GPR association rules.
Retrieving enzyme cuts targeting both bacteria for therapeutic action in human
To derive the sets of MCSs in common to both bacteria, reactions in MCSs were summed together, and MCSs from both models which had the same total mass balance equation were kept. The sets of MCSs in common were then converted into the corresponding minimal sets of genes with ASP logic programming, using GPRs derived from each bacterial model.
Protein structure models were retrieved using the AlphaFold entries53 associated to UniProt genes, and structure alignment was performed using algorithm FATCAT 2.077. All retrieved genes using the model’s GPRs could be associated to UniProt entries, but very few of these entries had known crystallized structures in PDB. As such, only AlphaFold entries were compared together.
Enzyme targets with good interspecies protein structure alignments were tested for existence of human homologues in the UniProt database using their E.C. Number. For enzymes with no human homologues, enzymes were kept as possible targets. For enzymes which had human homologues, the corresponding AlphaFold structure predictions were retrieved, and further protein structure alignments were performed to check druggability.
All MCSs with common enzymatic function between the two bacteria were reported in Supplementary Dataset 2 as well as their FATCAT alignment scores. We further detail the procedure for protein structure alignments in the next two subsections.
Interspecies protein structure alignments of enzyme targets
Protein structures with a FATCAT alignment RMSD above 3 Ångström were considered dissimilar between species (Supplementary Fig. 5). Excluded enzymes comprised Glutamyl-tRNA synthetase and reductase, and in particular, 3-dehydroquinate dehydratase, for which we confirmed through UniProt and InterPro that between P. aeruginosa and S. aureus, the enzymes had very different protein domains. Overall, we found that this threshold for scores of alignment between AlphaFold structure predictions was a helpful indicator of whether or not the proteins were similar between species.
Many MCSs would equivalently include exchange reactions or transporter reactions. Thus, we called these MCSs “growth medium dependent”. These are possible drug targets, but only in auxotrophic conditions, when amino acids are depleted from the medium. Although these cut sets might be challenging to use as therapeutic targets, we found them to be at least informative because if only the medium is depleted, then the enzymes become essential, and a drug targeting them would have an effect. These are also indicators of the main enzymes relating to a particular amino acid metabolism.
Estimating quality of enzyme targets for therapeutic applications to humans
FATCAT alignments between the S. aureus and P. aeruginosa structures and all human homologues were performed. For each alignment, three categories were considered, based on FATCAT scores: “Structurally equivalent”: (RMSD < 3) and (score < 10−6), “Structurally similar”: (3 ≤ RMSD < 5) and (10−6≤ score < 10−3), “Structurally dissimilar”: (RMSD≥5) and (score ≥ 10−3). Then, only enzymes with strictly less than 50% human homologues which had “Structurally equivalent” alignments with both bacteria were kept as possible targets (Supplementary Fig. 5).
Most of the enzymes with human homologues were eliminated through this procedure, the analysis excluded thirteen potentially not targetable enzymes out of sixteen. Almost all excluded enzymes, for which alignments were classified as “Structurally equivalent”, had identical InterPro protein domains between human and bacteria, despite the large phylogenetic distance, thus making for dangerous therapeutic targets.
Finally, we decided to exclude the target nucleoside diphosphate kinase (ATP:UDP) (36.73% “Structurally equivalent”, 18 proteins similar out of 49 homologues), even though it scored high in terms of interspecies protein structure alignment, as we believe targeting this enzyme would not be viable in human cells. As a result, only two enzymes with human homologues were kept at the end of the analysis (Supplementary Fig. 2).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated and analysed during the current study are provided in Supplementary Datasets and available from the corresponding author on request.
Code availability
Code for the analysis is provided at https://github.com/maxm4/paSAmcs/.
References
Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Maliniak, M. L., Stecenko, A. A. & McCarty, N. A. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: a single-center study. J. Cyst. Fibros. 15, 350–356 (2016).
Ahlgren, H. G. et al. Clinical outcomes associated with Staphylococcus aureus and Pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm. Med. 15, 67 (2015).
DeLeon, S. et al. Synergistic interactions of pseudomonas aeruginosa and staphylococcus aureus in an in vitro wound model. Infect. Immun. 82, 4718–4728 (2014).
Hotterbeekx, A., Kumar-Singh, S., Goossens, H. & Malhotra-Kumar, S. In vivo and In vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 7, 106 (2017).
Dalton, T. et al. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6, e27317 (2011).
Sibley, C. D. et al. Discerning the complexity of community interactions using a drosophila model of polymicrobial infections. PLoS Pathogens. 4, e1000184 (2008).
Woods, P. W., Haynes, Z. M., Mina, E. G. & Marques, C. N. H. Maintenance of S. aureus in co-culture with P. aeruginosa while growing as biofilms. Front. Microbiol. 9, 3291 (2018).
Alves, P. M. et al. Interaction between Staphylococcus aureus and Pseudomonas aeruginosa is beneficial for colonisation and pathogenicity in a mixed biofilm. Pathog. Dis. 76, fty003 (2018).
Phalak, P., Chen, J., Carlson, R. P. & Henson, M. A. Metabolic modeling of a chronic wound biofilm consortium predicts spatial partitioning of bacterial species. BMC Syst. Biol. 10, 90 (2016).
Phalak, P. & Henson, M. A. Metabolic modelling of chronic wound microbiota predicts mutualistic interactions that drive community composition. J. Appl. Microbiol. 127, 1576–1593 (2019).
Palsson, B. Ø. Systems biology: constraint-based reconstruction and analysis (Cambridge University Press, 2015).
Orth, J. D., Thiele, I. & Palsson, B. Ø. What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010).
Beck, A. E., Hunt, K. A. & Carlson, R. P. Measuring cellular biomass composition for computational biology applications. Processes 6, 38 (2018).
Lewis, N. E., Nagarajan, H. & Palsson, B. O. Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 10, 291–305 (2012).
Schuster, S., Dandekar, T. & Fell, D. A. Detection of elementary modes in biochemical networks: a promising tool for pathway analysis and metabolic engineering. Trends Biotechnol. 17, 53–60 (1999).
Acuña, V. et al. Modes and cuts in metabolic networks: complexity and algorithms. Biosystems 95, 51 – 60 (2009).
Hunt, K. A., Folsom, J. P., Taffs, R. L. & Carlson, R. P. Complete enumeration of elementary flux modes through scalable demand-based subnetwork definition. Bioinformatics 30, 1569–1578 (2014).
Jungreuthmayer, C., Ruckerbauer, D. E. & Zanghellini, J. regEfmtool: speeding up elementary flux mode calculation using transcriptional regulatory rules in the form of three-state logic. Biosystems 113, 37 – 39 (2013).
Gerstl, M. P., Jungreuthmayer, C. & Zanghellini, J. tEFMA: computing thermodynamically feasible elementary flux modes in metabolic networks. Bioinformatics 31, 2232–2234 (2015).
Pfeiffer, T., Sanchez-Valdenebro, I., Nuno, J. C., Montero, F. & Schuster, S. METATOOL: for studying metabolic networks. Bioinformatics 15, 251–257 (1999).
Gagneur, J. & Klamt, S. Computation of elementary modes: a unifying framework and the new binary approach. BMC Bioinform. 5, 175 (2004).
Price, N. D., Papin, J. A., Schilling, C. H. & Palsson, B. O. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 21, 162–169 (2003).
O’Brien, E. J., Lerman, J. A., Chang, R. L., Hyduke, D. R. & Palsson, B. Ø. Genome-scale models of metabolism and gene expression extend and refine growth phenotype prediction. Mol. Syst. Biol. 9, 693 (2013).
Joyce, A. R. & Palsson, B. Ø. Predicting gene essentiality using genome-scale in silico models. In Microbial Gene Essentiality: Protocols and Bioinformatics, Methods Mol. Biol. 416, 433–457 (2008).
Xavier, J. C., Patil, K. R. & Rocha, I. Metabolic models and gene essentiality data reveal essential and conserved metabolism in prokaryotes. PLoS Comput. Biol. 14, e1006556 (2018).
Folger, O. et al. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 7, 501 (2011).
Apaolaza, I. et al. An in-silico approach to predict and exploit synthetic lethality in cancer metabolism. Nat. Commun. 8, 459 (2017).
Blazier, A. S. & Papin, J. A. Reconciling high-throughput gene essentiality data with metabolic network reconstructions. PLOS Comput. Biol. 15, e1006507 (2019).
Suthers, P. F., Zomorrodi, A. & Maranas, C. D. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol. Syst. Biol. 5, 301 (2009).
Pratapa, A., Balachandran, S. & Raman, K. Fast-SL: an efficient algorithm to identify synthetic lethal sets in metabolic networks. Bioinformatics 31, 3299–3305 (2015).
Dehghan Manshadi, M., Setoodeh, P. & Zare, H. Rapid-SL identifies synthetic lethal sets with an arbitrary cardinality. Sci. Rep. 12, 14022 (2022).
Hädicke, O. & Klamt, S. Computing complex metabolic intervention strategies using constrained minimal cut sets. Metab. Eng. 13, 204–213 (2011).
Klamt, S. & Gilles, E. D. Minimal cut sets in biochemical reaction networks. Bioinformatics 20, 226–234 (2004).
Klamt, S. & Stelling, J. Combinatorial complexity of pathway analysis in metabolic networks. Mol. Biol. Rep. 29, 233–236 (2002).
Ballerstein, K., von Kamp, A., Klamt, S. & Haus, U.-U. Minimal cut sets in a metabolic network are elementary modes in a dual network. Bioinformatics 28, 381–387 (2012).
de Figueiredo, L. F. et al. Computing the shortest elementary flux modes in genome-scale metabolic networks. Bioinformatics 25, 3158–3165 (2009).
von Kamp, A. & Klamt, S. Enumeration of smallest intervention strategies in genome-scale metabolic networks. PLOS Comput. Biol. 10, 1–13 (2014).
Machado, D., Herrgård, M. J. & Rocha, I. Stoichiometric representation of gene-protein-reaction associations leverages constraint-based analysis from reaction to gene-level phenotype prediction. PLOS Comput. Biol. 12, e1005140 (2016).
Schneider, P., Kamp, A. V. & Klamt, S. An extended and generalized framework for the calculation of metabolic intervention strategies based on minimal cut sets. PLOS Comput. Biol. 16, e1008110 (2020).
Mahout, M., Carlson, R. P. & Peres, S. Answer set programming for computing constraints-based elementary flux modes: application to escherichia coli core metabolism. Processes 8, 1649 (2020).
Terzer, M. & Stelling, J. Large-scale computation of elementary flux modes with bit pattern trees. Bioinformatics 24, 2229–2235 (2008).
Thiele, S., von Kamp, A., Bekiaris, P. S., Schneider, P. & Klamt, S. CNApy: a CellNetAnalyzer GUI in Python for analyzing and designing metabolic networks. Bioinformatics 38, 1467–1469 (2022).
Vieira, V. & Rocha, M. CoBAMP: a Python framework for metabolic pathway analysis in constraint-based models. Bioinformatics 35, 5361–5362 (2019).
Seif, Y. et al. A computational knowledge-base elucidates the response of Staphylococcus aureus to different media types. PLOS Comput. Biol. 15, e1006644 (2019).
Poudel, S. et al. Revealing 29 sets of independently modulated genes in Staphylococcus aureus, their regulators, and role in key physiological response. Proc. Natl. Acad. Sci. 117, 17228–17239 (2020).
Renz, A. & Dräger, A. Curating and comparing 114 strain-specific genome-scale metabolic models of Staphylococcus aureus. NPJ Syst. Biol. Appl. 7, 30 (2021).
Bartell, J. A. et al. Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat. Commun. 8, 14631 (2017).
Yung, Y. P. et al. Reverse diauxie phenotype in Pseudomonas aeruginosa biofilm revealed by exometabolomics and label-free proteomics. NPJ Biofilms Microbiomes 5, 31 (2019).
McGill, S. L. et al. Pseudomonas aeruginosa reverse diauxie is a multidimensional, optimized, resource utilization strategy. Sci. Rep. 11, 1457 (2021).
Eadsforth, T. C. et al. Assessment of Pseudomonas aeruginosa N5,N10-Methylenetetrahydrofolate dehydrogenase—cyclohydrolase as a potential antibacterial drug target. PLoS One 7, e35973 (2012).
Stefaniak, J., Nowak, M. G., Wojciechowski, M., Milewski, S. & Skwarecki, A. S. Inhibitors of glucosamine-6-phosphate synthase as potential antimicrobials or antidiabetics-synthesis and properties. J. Enzyme Inhib. Med. Chem. 37, 1928–1956 (2022).
Varadi, M. et al. AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2022).
Nielsen, L. N. et al. Glyphosate has limited short-term effects on commensal bacterial community composition in the gut environment due to sufficient aromatic amino acid levels. Environ. Pollut. 233, 364–376 (2018).
Amorim Franco, T. M. & Blanchard, J. S. Bacterial branched-chain amino acid biosynthesis: structures, mechanisms, and drugability. Biochemistry 56, 5849–5865 (2017).
Gebser, M. et al. Theory Solving Made Easy with Clingo 5. In Technical Communications of the 32nd International Conference on Logic Programming (ICLP 2016), OpenAccess Series in Informatics (OASIcs) 52, 2:1–2:15 (2016).
Meylan, S. et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem. Biol. 24, 195–206 (2017).
Meylan, S., Andrews, I. W. & Collins, J. J. Targeting antibiotic tolerance, pathogen by pathogen. Cell 172, 1228–1238 (2018).
Hunt, K. A., Jennings, R. M., Inskeep, W. P. & Carlson, R. P. Stoichiometric modelling of assimilatory and dissimilatory biomass utilisation in a microbial community. Environ. Microbiol. 18, 4946–4960 (2016).
Hunt, K. A., Jennings, R. M., Inskeep, W. P. & Carlson, R. P. Multiscale analysis of autotroph-heterotroph interactions in a high-temperature microbial community. PLOS Comput. Biol. 14, e1006431 (2018).
Zelezniak, A. et al. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc. Natl. Acad. Sci. 112, 6449–6454 (2015).
Paczia, N. et al. Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 11, 122 (2012).
Giri, S. et al. Metabolic dissimilarity determines the establishment of cross-feeding interactions in bacteria. Curr. Biol. 31, 5547–5557.e6 (2021).
Magnúsdóttir, S. et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nat. Biotechnol. 35, 81–89 (2017).
Thiele, I. et al. Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol. Syst. Biol. 16, e8982 (2020).
Oberhardt, M. A., Puchałka, J., Fryer, K. E., Martins dos Santos, V. A. P. & Papin, J. A. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J. Bacteriol. 190, 2790–2803 (2008).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Schneider, P., Bekiaris, P. S., von Kamp, A. & Klamt, S. StrainDesign: a comprehensive Python package for computational design of metabolic networks. Bioinformatics 38, 4981–4983 (2022).
Halsey, C. R. et al. Amino acid catabolism in staphylococcus aureus and the function of carbon catabolite repression. mBio 8, e01434–16 (2017).
Lieven, C. et al. MEMOTE for standardized genome-scale metabolic model testing. Nat. Biotechnol. 38, 272–276 (2020).
Ebrahim, A., Lerman, J. A., Palsson, B. O. & Hyduke, D. R. COBRApy: COnstraints-based reconstruction and analysis for Python. BMC Syst. Biol. 7, 74 (2013).
Janhunen, T. et al. Clingo goes linear constraints over reals and integers. Theory Pract. Logic Progr. 17, 872–888 (2017).
Pey, J. & Planes, F. J. Direct calculation of elementary flux modes satisfying several biological constraints in genome-scale metabolic networks. Bioinformatics 30, 2197–2203 (2014).
Ray, O., Soh, T. & Inoue, K. Analyzing pathways using ASP-based approaches. In Algebraic and Numeric Biology, Lecture Notes in Computer Science 6479, 167–183 (2012).
Frioux, C., Schaub, T., Schellhorn, S., Siegel, A. & Wanko, P. Hybrid metabolic network completion. In Logic programming and nonmonotonic reasoning. Lecture Notes in Computer Science 10377, 308–321 (2017).
Lierler, Y. What is answer set programming to propositional satisfiability. Constraints 22, 307–337 (2017).
Li, Z., Jaroszewski, L., Iyer, M., Sedova, M. & Godzik, A. FATCAT 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 48, W60–W64 (2020).
Acknowledgements
M.M. and S.P. were supported by ITMO Cancer of Aviesan within the framework of the 2021-2030 Cancer Control Strategy, on funds administered by Inserm. Réf. N∘21CM141-00. R.P.C. was supported by National Institutes of Health award U01EB019416.
Author information
Authors and Affiliations
Contributions
M.M., R.C., L.S., and S.P. designed the study. R.C. and S.P. lead the study. M.M. and R.C. contributed to the paper’s figures. M.M. developed the software for the paper with supervision from L.S. and S.P.; M.M. performed the experiments and wrote the paper with input from all authors. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mahout, M., Carlson, R.P., Simon, L. et al. Logic programming-based Minimal Cut Sets reveal consortium-level therapeutic targets for chronic wound infections. npj Syst Biol Appl 10, 34 (2024). https://doi.org/10.1038/s41540-024-00360-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41540-024-00360-6