Abstract
N6,2′-O-dimethyladenosine (m6Am), a terminal modification adjacent to the mRNA cap, is a newly discovered reversible RNA modification. Yet, a specific and sensitive tool to directly map transcriptome-wide m6Am is lacking. Here, we report m6Am-seq, based on selective in vitro demethylation and RNA immunoprecipitation. m6Am-seq directly distinguishes m6Am and 5′-UTR N6-methyladenosine (m6A) and enables the identification of m6Am at single-base resolution and 5′-UTR m6A in the human transcriptome. Using m6Am-seq, we also find that m6Am and 5′-UTR m6A respond dynamically to stimuli, and identify key functional methylation sites that may facilitate cellular stress response. Collectively, m6Am-seq reveals the high-confidence m6Am and 5′-UTR m6A methylome and provides a robust tool for functional studies of the two epitranscriptomic marks.
Similar content being viewed by others
Introduction
More than 160 different chemical modifications have been found so far1. N6-methyladenosine (m6A) is the most abundant internal modification on mRNA and lncRNA in eukaryotes2,3,4. In addition to m6A, there exists another reversible modification in higher eukaryotes, called N6, 2′-O-dimethyladenosine (m6Am), which is precisely located at the first transcribed nucleotide and hence adjacent to the cap structure of mRNA5,6. A total of 50–80% of adenosine-starting mammalian mRNAs are believed to be m6Am modified6. It is catalyzed by PCIF17,8,9,10, a protein that interacts with the phosphorylated CTD of RNAPII11, and could be removed by the m6A demethylase FTO12,13. Thus, m6Am is dynamically regulated by FTO and PCIF1, leading to the direction of the cap epitranscriptomics.
The development of transcriptome-wide sequencing technologies for various RNA modifications has greatly facilitated the field of epitranscriptomics14. The most widely used method for m6Am/m6A mapping relies on m6A antibodies, which do not discriminate between the two functionally distinct modifications15,16. While m6A is enriched around the stop codon, it is challenging to distinguish m6Am from 5′-UTR m6A, which constitutes a significant part of m6A methylome and has been shown to be functionally important in various biological processes17,18,19. There have been efforts aiming to distinguish the two modifications8,9,10,20,21; yet they suffer from the low efficiency of UV crosslinking, the inaccuracy of TSS annotation, and the limited activity of 5′ exonuclease, thus compromising the sensitivity and precision of m6Am detection. While the use of PCIF1 knockout (KO) cell lines has improved the confidence of m6Am detection9,10, it is an indirect approach and is incompatible for epitranscriptome analysis in human tissues and biological samples where genetic manipulation is challenging. Thus, a sensitive and direct method for transcriptome-wide m6Am identification is highly desired.
Here, we develop m6Am-seq to investigate the prevalence, topology, and dynamics of m6Am in the human transcriptome. m6Am-seq relies on an in vitro demethylation reaction that selectively removes m6Am while keeps m6A intact, thereby discriminating genuine m6Am at the mRNA cap and 5′-UTR m6A. Using m6Am-seq, we identify 2166 m6Am sites at base resolution from 1652 peaks and 1307 5′-UTR m6A peaks throughout the human transcriptome. Moreover, we show that m6Am and 5′-UTR m6A respond dynamically to heat shock and hypoxia conditions, and identify hundreds of stress-induced m6Am and 5′-UTR m6A peaks that could mediate adaptive response to the stressors. Altogether, we provide a tool to directly and selectively distinguish and profile m6Am and 5′-UTR m6A; we anticipate that m6Am-seq will pave the way for future functional studies of m6Am in various biological systems.
Results
A selective demethylation reaction to distinguish m6Am and m6A
m6Am-seq utilizes a selective demethylation reaction to achieve specific and sensitive detection of m6Am. While both m6Am and m6A are recognized by antibodies, we envisioned that the unique cap-adjacent structural context of m6Am could be selectively read out in vitro by demethylase (Supplementary Fig. 1a). To this end, we tested ALKBH5 and FTO, two known RNA demethylases22,23. We found that ALKBH5 does not act on m6Am, consistent with previous biochemical data13. However, it does not demethylate m6A to completion (~80%, Supplementary Fig. 1b), complicating subsequent analysis. In contrast, FTO prefers m6Am but demethylates both modifications with high efficiency (>98% and ~60% for m6Am and m6A, respectively), under recommended conditions from literature13 (Supplementary Fig. 1c).
Neither of the demethylation reactions would be good enough to distinguish m6Am and m6A in their present forms. The ideal condition would remove one modification to completion while leaving another intact. FTO and ALKBH5 are Fe(II)/2-KG-dependent dioxygenases, whose in vitro demethylation activity is promoted by l-ascorbic acid22,23. As a reducing agent, it is capable of reducing redox-active metals such as iron, thereby increasing the pro-oxidant chemistry of these metals24. We thus hypothesized that by omitting l-ascorbic acid from our conditions, we might be able to modulate the in vitro activity of FTO so that it only acts on the preferred, cap-adjacent m6Am but not the internal m6A. To our delight, after removing l-ascorbic acid and further optimizing the demethylation conditions, >95% of m6Am was removed without significantly affecting the m6A level (Fig. 1c). We further verified the selectivity of our demethylation condition using a dually modified probe containing both cap m6Am and internal m6A, which were 76 nt apart from each other (Supplementary Fig. 1d). Thus, we identified a demethylation reaction that is specific for m6Am.
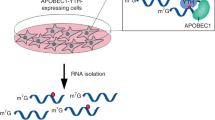
a Scheme of m6Am-seq. Total RNA (>200 nt) was fragmented (input) and immunoprecipitated with cap-m7G antibody (m7G-IP). The cap structure-containing RNA was subjected to [FTO (+)] or [FTO (−)] treatment. The [FTO (+)] or [FTO (−)] samples were immunoprecipitated with an m6A antibody and sequenced for m6Am detection. The sequencing profiles of the [FTO (+) m6A-IP] and [FTO (−) m6A-IP] samples were normalized and compared. The “Demethylase-sensitive peaks” and “Demethylase-insensitive peaks” were identified. The demethylase-sensitive peaks bear the m6Am, while the demethylase-insensitive peaks bear the m6A (Online Methods). b The m6Am and m6A levels of input, m7G-IP, and flowthrough samples were quantified by LC–MS/MS, respectively. m6Am was significantly enriched (>100 fold) by cap-m7G antibody while m6A level decreased mildly. Values represent mean ± SD (n = 3 independent samples in “Input”, “IP” and “Flowthrough” respectively). c The m6Am and m6A levels of [FTO (+)] and [FTO (−)] samples were quantified by LC–MS/MS, respectively. >95% m6Am was demethylated and m6A was negligibly altered under optimized demethylation conditions. The chromatograms of U (black) and m6Am (red) are scaled to the left Y axis, and the chromatograms of m6A (blue) are scaled to the right Y axis. Source data are provided as a Source Data file.
m6Am-seq directly detects m6Am
To map the m6Am in the human transcriptome, we developed m6Am-seq, which combines the selective demethylation reaction with RNA immunoprecipitation (Fig. 1). Two key RNA immunoprecipitation procedures are utilized, which are a cap-m7G IP and an m6A IP step. Because the global m6A level is around 20-fold higher than that of m6Am in total RNA (Supplementary Fig. 1e), and the vast majority of m6Am is localized in mRNA cap (Supplementary Fig. 1f), we used the cap-m7G IP step to enrich for m6Am and deplete m6A that is mostly found in the 3′-UTR. As expected, immunoprecipitation with a cap-m7G antibody for fragmented RNA resulted in more than 100-fold enrichment of m6Am and a simultaneous 2-fold reduction of m6A (Fig. 1b). Moreover, ~90% of peaks are located at and near 5′-UTR, suggesting 5′-end of mRNAs were successfully and specifically enriched using size selected total RNA (>200 nt) as input, without the prerequisite to purify mRNA (Supplementary Fig. 2a).
The immunoprecipitated RNA was then subjected to the selective demethylation reaction or mock treatment, which was followed by the second IP step utilizing an m6A antibody (Fig. 1a). The FTO (−) sample would contain both m6Am and 5′-UTR m6A signals, while the FTO (+) sample would contain only m6A. By comparing the methylome profiles of the two samples, we were able to detect m6Am, which is sensitive to demethylase treatment, from 5′-UTR m6A, which is insensitive and conserved in the two samples. Indeed, we observed a bimodal-like distribution of fold change of peak intensity: peaks on the right side experienced reduced intensity upon demethylase treatment, suggesting they are demethylase-sensitive m6Am modification; while peaks on the left were not significantly altered or even slightly increased, suggesting they are retained 5′-UTR m6A peaks (Supplementary Fig. 2b, c). Thus, m6Am-seq selectively distinguishes the two modifications.
m6Am-seq detects transcriptome-wide m6Am and 5′-UTR m6A
We next applied m6Am-seq to characterize m6Am and 5′-UTR m6A methylome in HEK293T cells. We found that m6Am-seq results are highly correlated between biological replicates (correlation coefficient = 0.9821, Supplementary Fig. 2d), demonstrating its robustness. We identified 1652 and 1307 high-confidence m6Am and 5′-UTR m6A peaks from 1635 and 1297 genes, exhibiting the characteristic BCA (B = C/U/G) and GGACH (H = A/C/U) motif, respectively (Fig. 2a, b and Supplementary Data 1, 2). m6Am shows the highest enrichment at annotated TSSs, while the summit of m6A is clearly within 5′-UTR; the peak density of m6Am is also 3-fold higher than that of 5′-UTR m6A, demonstrating that m6Am is more concentrated than m6A around the TSSs (Supplementary Fig. 3a, b). Representative methylation profiles are shown in Fig. 2c. In one example, the ATF5 gene has multiple isoforms with distinct TSS and alternative first exons; we observed m6Am modification for one transcript and 5′-UTR m6A for another transcript (Fig. 2c), suggesting differential cis-regulatory elements in regulating the epitranscriptomic status of different transcripts from the same gene. Gene ontology (GO) terms analysis showed that transcripts containing m6Am and 5′-UTR m6A are most strongly enriched in cell cycle and RNA metabolism, respectively, suggesting their differential functions (Supplementary Fig. 3c, d).
Motif analysis of m6Am peaks (a) and 5′-UTR m6A peaks (b) identified by m6Am-seq. c Representative views of typical m6Am peaks and 5′-UTR m6A peaks on mRNA. The methylation peak in the 5′-UTR of PAX6 was lost in “FTO (+) m6A-IP” samples, suggesting a demethylase-sensitive m6Am modification (top); the methylation peak in the 5′-UTR of SQLE retained well in “FTO (+) m6A-IP” samples, suggesting a demethylase-insensitive m6A modification (middle); while multiple isoforms of ATF5 were differentially marked by m6Am and 5′-UTR m6A (bottom). Pink and blue background colors denote m6Am and m6A signals, respectively. d Venn diagram showing the overlap between m6Am peaks identified directly by m6Am-seq and m6Am peaks identified indirectly by m6A IP using PCIF1-KO samples. e Boxplot showing that the peak intensity of m6Am (red, n = 1652), but not that of m6A (blue, n = 1307), was significantly reduced upon PCIF1 depletion. Statistical significance of the difference was determined by unpaired two-sided Mann–Whitney U-test. ****P < 2.2e−16. Boxes represent 25th–75th percentile (line at median) with whiskers at 1.5*IQR. Source data are provided as a Source Data file.
To further demonstrate the accuracy of m6Am-seq, we applied standard m6A IP experiments to PCIF1-KO HEK293T cells. Based on the reduction of signals compared to WT cells, we detected 1357 PCIF1-dependent peaks; notably, 1200 out of the 1357 peaks were identified by m6Am-seq (Fig. 2d and Supplementary Fig. 3e), suggesting the high confidence of m6Am-seq. The remaining peaks all exhibited m6Am-like property (i.e., sensitive to demethylation) in m6Am-seq but were not called due to the stringent cutoff we used. On the other hand, 5′-UTR m6A peaks identified by m6Am-seq were not affected upon PCIF1 depletion (Fig. 2e and Supplementary Fig. 3f), further demonstrating the specificity of m6Am-seq. Taken together, these results prove that m6Am-seq identifies m6Am with high confidence. It is also worth mentioning that m6Am-seq is applicable for m6Am detection in tissues or cell populations where PCIF1 KD or KO is not feasible.
Comparisons of existing m6Am detection methods
We then compared m6Am-seq results with m6Am-Exo-seq and miCLIP9,10. Under normalized sequencing depth, the averaged peak coverage of m6Am-seq (~387) was ~4× and 21× higher than that of m6Am-Exo-seq (~95) and miCLIP (~18) (Fig. 3a). In addition, m6Am peaks by m6Am-seq exhibit a strong tendency towards the annotated TSS, in comparison to a modest enrichment within 5′-UTR by miCLIP and m6Am-Exo-Seq (Fig. 3b). Thus, m6Am-seq outperforms existing methods in m6Am detection.
a Boxplot showing that the normalized peak coverage by m6Am-seq (red, n = 1652), m6Am-Exo-seq (blue, n = 3169), and miCLIP (yellow, n = 2217). Statistical significance of the difference was determined by unpaired two-sided Mann–Whitney U-test. ****P < 2.2e−16 for m6Am-seq vs m6Am-Exo-seq, m6Am-seq vs miCLIP. Boxes represent 25th–75th percentile (line at the median) with whiskers at 1.5*IQR. b Metagene profiles of m6Am distribution identified by the three sequencing methods. Each segment was normalized according to its average length in RefSeq annotation. c Venn diagram showing the overlap of m6Am-marked genes by m6Am-seq and by miCLIP. d Boxplot showing the normalized peak coverage of shared m6Am peaks (yellow, n = 456, identified in both m6Am-seq and miCLIP) and m6Am-seq-unique m6Am peaks (pink, n = 1179) in the m6Am-seq and miCLIP data sets, respectively. Boxes represent 25th–75th percentile (line at the median) with whiskers at 1.5*IQR. e An example of m6Am (in the RBM48 gene), which was missed by miCLIP due to its limited sensitivity but is clearly identified by m6Am-seq. The m6Am peak is lost in PCIF1-KO data sets, further proving the detection confidence of m6Am-seq. Pink background color denotes m6Am signal. f An example of mis-annotated m6Am (in the ABCD1 gene) by miCLIP. This peak is not affected by FTO treatment nor PCIF1-KO, showing that it is a 5′-UTR m6A modification. Blue background color denotes m6A signal. Source data are provided as a Source Data file.
Because m6Am-seq and miCLIP can both detect m6Am and m6A, we performed comparisons of the two methods10,20 (Fig. 3c and Supplementary Fig. 4a, b). We used the recent miCLIP data relying on PCIF1-KO for detailed comparison10. Approximately 21% (456/2217) of m6Am marked genes by miCLIP overlap with that of m6Am-seq (Fig. 3c). We found that miCLIP data sets showed very low coverage, providing an explanation for the limited sensitivity of miCLIP (Fig. 3d, e). While it is possible that the stringent cutoff of m6Am-seq may favor specificity over sensitivity, a significant portion of miCLIP-identified m6Am peaks could be mis-annotated and are instead 5′-UTR m6A peaks (Fig. 3f).
It was proposed that m6Am controls mRNA stability12, based on the m6Am methylome identified by miCLIP. Given the observation that miCLIP may mis-annotate the two modifications, we compared the expression of m6Am-containing genes, which are identified by m6Am-seq, in wild-type and PCIF1-KO cells. Different from the previous conclusion12, we found that the loss of m6Am did not alter the expression of their host mRNA (Supplementary Fig. 4c).
m6Am-seq detects m6Am at single-base resolution
Because m6Am is the first transcribed nucleotide, one would expect to identify m6Am at base resolution by searching for A-starting transcripts (i.e., those showing sudden drop-off of sequencing reads at the 5′ end). However, even when we used a library preparation procedure that preserves the first transcribed nucleotide, we did not observe an enrichment of A nucleotide in the m6Am/m6A enriched FTO (−) data set (Fig. 4a), suggesting that its TSS location alone is not sufficient to detect m6Am sites. Thus, we defined a “start rate difference” score (SRD score) for each nucleotide to quantify sequencing reads beginning from it (see “Methods” section). Application of these metrics immediately resulted in accurate identification of the m6Am site in synthetic spike-in RNAs (Supplementary Fig. 5a) and ~95.8% of A nucleotides in the m6Am/m6A enriched FTO (−) data set (Fig. 4a). In total, we identified 2166 m6Am sites from 1459 genes (Supplementary Data 3). For instance, mRNA-coding gene PRR11 and a lncRNA LOC101929147 both have alternative TSSs that start with an A nucleotide, and we showed that all these transcripts are marked by m6Am (Fig. 4b and Supplementary Fig. 5b). In addition, while m6Am is associated with a BCA motif, our single-base m6Am methylome revealed ~34% non-BCA m6Am sites (Supplementary Fig. 6a). We found also a weaker signal of GRO-seq and H3K27ac ChIP-seq for the non-BCA m6Am sites, but no difference in H3K4me3, H3K4me1, and H3K9me3 signals (Supplementary Fig. 6b–f). Altogether, these data indicate that m6Am-seq enables m6Am detection at single-base resolution.
a Nucleotide proportion of TSS identified by the sudden drop-off of sequencing reads (left) and by “start rate difference” (SRD) metrics. b A representative view of three single m6Am sites on the transcripts of PRR11, which were all supported by CAGE data. Three adenosine residues with a high SRD score (red bar) were defined as m6Am sites, which overlapped exactly with CAGE sites (green triangles). Each segment was normalized according to its average length in RefSeq annotation. Pink background color denotes m6Am signal. c A metaplot showing that the vast majority of m6Am sites by m6Am-seq overlapped with CAGE sites, suggesting that these m6Am sites are indeed TSSs. Compared with miCLIP, m6Am sites identified by m6Am-seq were much closer to the CAGE TSSs. Two miCLIP data sets were used for comparison: an m6Am list reported in 2015 20 and an updated m6Am list in 2019 10, based on PCIF1-KO. Source data are provided as a Source Data file.
To further characterize the accuracy of the identified m6Am sites, we performed the cap-analysis of gene expression (CAGE) experiments and compared the m6Am sites with annotated and CAGE-identified TSSs. We found that most of the m6Am sites are present downstream of the RefSeq-annotated TSSs, with only 14% of m6Am sites mapped at the annotated TSSs (Supplementary Fig. 7a). This observation suggested that existing methods based on annotated TSSs information can be inaccurate for m6Am mapping. For instance, the m6Am site of TMET241 would have been located internally within 5′-UTR if the annotated TSSs were used (Supplementary Fig. 7b); and two m6Am peaks are clearly identified for RAB5A, which has only one annotated TSS (Supplementary Fig. 7c). We next plotted the distance from m6Am sites to their nearest CAGE TSS sites, and observed that the vast majority of m6Am sites overlapped with CAGE TSSs (Fig. 4c), suggesting that these m6Am sites are indeed TSSs. Compared with miCLIP10,20, m6Am sites by m6Am-seq were much closer to the CAGE TSSs (Fig. 4c). These data highlight the reliability of m6Am-seq in obtaining high-confidence m6Am sites.
Heat shock induces dynamic m6Am and 5′-UTR m6A methylations
Stress-induced RNA modification is an important feature of dynamic epitranscriptomic regulation under physiological conditions15,25,26,27. For instance, it has been shown that in response to heat shock stress, m6A deposited to the 5′-UTR of newly transcribed mRNAs promotes cap-independent translation initiation, providing a mechanism of selective mRNA translation during general translation inhibition17,19. In light of this, we applied m6Am-seq to characterize the dynamic m6Am and 5′-UTR m6A methylome under heat shock conditions. We identified 45 upregulated m6Am peaks and 90 5′-UTR m6A peaks, as well as 36 and 67 downregulated m6Am and 5′-UTR m6A peaks (Fig. 5a, b and Supplementary Fig. 8a, b and Supplementary Fig. 9a–d and Supplementary Data 4). Among them, we found key heat shock-associated transcripts with elevated 5′-UTR m6A: notable examples include HSPA1A, HSPA1B, and DNAJB1 (Supplementary Fig. 10a, b), whose transcripts undergo m6A-dependent but cap-independent selective translation17. These results by m6Am-seq accurately recapitulated heat shock-induced, functional 5′-UTR m6A methylation.
a–d Volcano plot showing stress-induced m6Am and 5′-UTR m6A: heat shock-inducible m6Am peaks (a) and 5′-UTR m6A peaks (b), and hypoxia-inducible m6Am peaks (c), and 5′-UTR m6A peaks (d). Red circles denote stress-upregulated peaks; blue circles denote stress-downregulated peaks. Representative views of elevated m6Am peaks in CHOP (e) and GLUT1 (f) under hypoxia stress. Representative views of elevated 5′-UTR m6A peaks in TP53 (g) and PTEN (h) under hypoxia stress. Pink and blue background colors denote m6Am and m6A signals, respectively.
Unexpectedly, a subset of transcripts previously proposed to rely on m6A-dependent translation under heat shock17 did not contain m6A modification in the 5′-UTR; instead, m6Am-seq reveals that they are dynamically modified by m6Am (Supplementary Fig. 10c). For instance, using a method that does not distinguish between m6Am and 5′-UTR m6A, upregulation of 5′-UTR m6A level in EGR1 was believed to mediate the stress-induced selective translation. However, using m6Am-seq, we found that EGR1 contained m6Am rather than 5′-UTR m6A (Supplementary Fig. 10d). Therefore, cap-independent translation of such transcripts including EGR1 is unlikely to be mediated by m6A. Whether or not m6Am, which is located at TSS and immediately after the m7G cap, can also promote stress-induced selective translation would be of interest for future studies. Overall, m6Am-seq provides accurate methylome maps to enable the study of the physiological roles of the two RNA modifications.
Dynamic m6Am and 5′-UTR m6A methylome under hypoxia conditions
We next determined whether m6Am and 5′-UTR m6A methylation can be dynamically regulated by hypoxia. We were able to identify 362 and 638 elevated m6Am and 5′-UTR m6A peaks respectively, with 31 and 63 decreased m6Am and 5′-UTR m6A peaks (Fig. 5c, d and Supplementary Fig. 8c, d and Supplementary Data 5). While some of the hypoxia-specific genes show inducible m6Am and 5′-UTR m6A modifications, many transcripts with unaltered expression levels are also dynamically modified (Supplementary Fig. 9e–h). Overall, we observed a more evident increase of m6Am and m6A methylome under hypoxia conditions than heat shock conditions.
To cope with hypoxia, mammals have evolved key adaptive mechanisms including cellular adaptations of protein synthesis, energy metabolism, mitochondrial respiration, lipid, and carbon metabolism as well as nutrient acquisition28. Given the dramatic change of m6Am level, we examined whether dynamic m6Am may participate in the adaptive mechanisms during the hypoxia condition. Intriguingly, our GO analysis showed that genes with elevated m6Am levels are linked to the endoplasmic reticulum (ER) stress-response regulation (Supplementary Fig. 11a). For instance, it has been documented that transcriptional and translational upregulation of both CHOP and ATF3 are important to activate the expression of downstream genes in response to stress29,30; in line with this, we observed significantly elevated m6Am levels in their mRNAs (Fig. 5e and Supplementary Data 5). In addition to ER stress, oxygen deprivation also reprograms intracellular metabolism31. For instance, hypoxia can enhance the mRNA and protein levels of HIF1 target gene GLUT1 to achieve glucose transport31; in agreement with this, we also observed increased m6Am level in the GLUT1 mRNA (Fig. 5f). Thus, we identified a dynamic m6Am program in key transcripts that are utilized by cells to adapt to hypoxia.
Distinct from the m6Am-marked genes, GO analysis of genes with elevated 5′-UTR m6A indicated an enrichment of the p53 signaling pathway (Supplementary Fig. 11b). In fact, hypoxia has frequently been described to be a p53 inducer32. Along this line, p53 and many of its downstream genes, such as MDM2, PTEN, PIDD1, etc., were found to contain elevated 5′-UTR m6A under hypoxia stress (Fig. 5g, h and Supplementary Data 5). Since induced 5′-UTR m6A by heat shock has been shown to enable cap-independent translation, it is possible that protein synthesis of the p53 signaling pathway could be modulated by the stress-induced m6A. Altogether, our results suggest that dynamic m6Am and m6A as key components of the adaptive response to hypoxia.
Discussion
More than 40 years have passed since the initial documentation of m6Am was made5,6. The recent discovery of its “eraser” and “writer” has led to a renewed interest in this reversible modification33. Yet its biological functions remain enigmatic, due to a lack of sensitive methods that detect the modification at the transcriptome-wide level. Here, we report m6Am-seq, which enables specific and robust profiling of m6Am in the human transcriptome.
The development of m6Am-seq is inspired by existing epigenomic and epitranscriptomic methods utilizing biologically important demethylases. For instance, TAB-Seq34 and TAPS35 rely on the TET proteins, which oxidize 5-methylcytosine (5mC), to detect 5mC and its oxidative derivatives in the genome. In DM-tRNA-seq36, ARM-seq37, and m1A-MAP38, the RNA/DNA demethylase AlkB has been used to map N1-methyladenosine (m1A) in tRNA and mRNA. In this study, we expand the available toolbox by repurposing FTO to detect m6Am and m6A, two dynamic and reversible epitranscriptomic marks. Different from the existing methods that use demethylases to indiscriminately remove all substrates, we manipulated the in vitro activity of FTO to selectively demethylate just one of the two cognate substrates. This is achieved by the discovery of a controlled in vitro demethylation reaction, which omits the key cofactor l-ascorbic acid from the biochemical condition. Owing to its capacity to reduce Fe3+ to the catalytically active Fe2+, l-ascorbic acid can enhance the in vitro demethylation activity24,39. By leaving out l-ascorbic acid from the in vitro reactions, FTO still possesses a high m6Am demethylation activity while minimally affects the level of m6A, hence allowing selective detection of m6Am.
Interestingly, the in vivo bioavailability of ascorbate, the dominant form of vitamin C under physiological pH conditions, has been demonstrated to impact the epigenome via TETs and JmjC domain-containing demethylases, ultimately leading to phenotypic changes in development, aging, cancer and other diseases24. It remains to be seen whether or not the deficiency of vitamin C might alter m6Am or m6A methylome and hence epitranscriptome reprogramming inside of cells.
Except for the selective demethylation condition omitting l-ascorbic acid, major steps of m6Am-seq are well-known meRIP protocols. Hence, m6Am-seq is straightforward and highly reproducible. Its high sensitivity and accuracy are also enabled by the combination of cap-m7G IP and m6A IP, which builds on the knowledge that m6Am is adjacent to the m7G cap. It should be noted that m6Am-seq does not detect internal m6Am, although the amount of the internal m6Am in mRNAs seems negligible. Only one internal m6Am site in U2 snRNA has been documented so far. In m6Am-seq, we used a demethylation reaction to distinguish m6Am from m6A, other selective chemistry could be exploited for the determination of additional modifications. For instance, dNTP-sensitivity during reverse transcription40 and peroxidate oxidation41 could be combined with cap-m7G IP to detect ribose methylation at the first and second transcribed nucleotides of mRNA. In addition, given the diversity of RNA cap structures42,43, including NAD+, NADH, desphospho-CoA, and 2,2,7-trimethylguanosine, tailored combination of IP enrichment and a discrimination procedure, as demonstrated in m6Am-seq, could have broad applications in analyzing the epitranscriptomic state of the transcriptome.
m6Am-seq distinguishes m6Am from m6A and provides a high-confidence m6Am methylome. While a previous study used miCLIP for m6Am identification and has indicated a role of m6Am in controlling mRNA stability12, this conclusion was disputed by more recent studies led by independent groups7,9,13. Using m6Am-seq, we corroborated the latter studies and found that m6Am does not affect mRNA stability. Because miCLIP does not separate well m6Am from m6A, it is likely that the observed effect in mRNA stability is caused by m6A13, which has been shown to increase mRNA stability when recognized by certain m6A readers44. Nevertheless, such disagreement underscores the importance of a specific method to accurately map m6Am for future functional studies.
We showed that m6Am is dynamically regulated by heat shock and hypoxia, hinting that m6Am is important for cellular stress response. While a specific role of m6Am remains to be elucidated, we found that many well-known stress-induced genes exhibiting elevated protein levels showed increased m6Am methylation levels upon stimuli. Such a positive correlation appears to be the opposite in normal human tissues21; nevertheless, this does not rule out the possibility of a unique role of m6Am under stress. Consistent with the positive association, ribosome profiling experiments also revealed a positive role of m6Am in translation7. Given that 5′-UTR m6A has been demonstrated to mediate cap-independent translation17,19, it is tempting to speculate that m6Am plays a role in stress-induced mRNA translation.
Whether or not m6Am may participate in cap-independent translation is an open question. It is known that the mRNA cap structure plays an important role in translation initiation. Akichika et al. showed that m6Am does not modulate the binding affinity of eIF4E to the cap structure, suggesting that other cap-binding factors could be involved7. For instance, it has been shown that eIF3 and ABCF1 can selectively recognize 5′-UTR m6A and mediate the cap-independent translation18,19. Thus, future identification of potential m6Am readers, particularly those in the integrated stress response, could help elucidate the detailed function and mechanism of m6Am.
In summary, we report a tool for transcriptome-wide mapping of m6Am and 5′-UTR m6A, and reveal the dynamic features of the two epitranscriptomic marks under stress conditions. We anticipate that m6Am-seq will lead to new territories of RNA biology and open opportunities in the field of “cap epitranscriptomics”.
Methods
Antibodies and cell culture
Rabbit anti-m6A antibody was purchased from EMD Millipore (ABE572). Anti-m7G-cap monoclonal antibody was obtained from MBL (RN016M). HEK293T cells were cultured at 37 °C in DMEM medium supplemented with 10% FBS, 1% GlutaMAX and 0.5% penicillin/streptomycin with 5% CO2. For heat shock, cells were grown to a confluence of 80% in 10-cm plastic dishes, placed in a 43 °C water bath for 2 h, and then recover for 6 h in normal growing conditions. For hypoxia treatment, cells were grown to 80%, carried out in Hypoxia Hood which was maintained at 37 °C, 5% CO2, 1% oxygen for 24 h.
RNA isolation
TRIzol was used for total RNA isolation according to the manufacturer’s protocol. <200 nt RNA was deleted using the MEGAclear Transcription Clean-Up kit (Ambion, AM1908).
Quantification of m6A and m6Am by LC–MS/MS
For m6Am, 150 ng RNA was decapped by 10 units of RppH (NEB, M0356S) in Thermopol buffer for 5 h at 37 °C. RNA was purified by RNA clean & concentrator kit (Zymo Research, R1015). Purified RNA was digested by nuclease P1 (Sigma, N8630) with 10 mM ammonium acetate at 42 °C for 6 h, then mixed with 50 mM MES buffer (pH 6.5) and 0.5 U Shrimp Alkaline Phosphatase (NEB, M0371S), and incubated at 37 °C for another 6 h. For m6A, 150 ng RNA was digested into single nucleosides by nuclease P1 with 10 mM ammonium acetate at 42 °C for 6 h, then incubated with 50 mM MES buffer (pH 6.5) and 0.5 U Shrimp Alkaline Phosphatase for another 6 h at 37 °C.
The digested nucleosides were separated by the ultra-performance liquid chromatography on C18 column, and detected by the triple-quadrupole mass spectrometer (AB SCIEX QTRAP 6500). The multiple reaction-monitoring (MRM) mode was monitored: m/z 268.0–136.0 (A), m/z 245.0–113.1 (U), m/z 282.0–150.1 (m6A), m/z 296.0–150.0 (m6Am). The concentrations of m6A and m6Am levels in RNA samples were calculated from the standard curves.
Synthesis of m6Am/Am and m6A probes
cap-m6Am/Am RNA: template was obtained by PCR, which contains the T7 promoter sequence and ~150 bp DNA sequence. Primers used to amplify the spike-in template was provided (Supplementary Data 6). In vitro transcription reaction contained: reaction buffer (ThermoFisher Scientific, AM1354), 800 ng template DNA, T7 Enzyme Mix (ThermoFisher Scientific, AM1354), 15 mM ATP/UTP/CTP/GTP, and 15 mM cap analogs (Trilink, N-7113 or N-7102, N-7102 was ordered via email addressed to orders@trilinkbiotech.com). The reaction was incubated overnight at 37 °C. After incubation, 1 μl of TURBO DNase (ThermoFisher Scientific, AM1354) was added to the reaction, and the reaction was incubated at 37 °C for 1 h. The RNA was purified by phenol–chloroform isolation and ethanol precipitation. The precise RNA was purified from 7 M urea/15% acrylamide gels using elution buffer (0.3 M sodium acetate, 1 mM EDTA, 0.05% SDS) at 37 °C for 12 h. Next, the RNA was precipitated with ethanol. The probes were used as spike-ins and for probe demethylation assay. The sequence of cap-m6Am/Am RNA probes was list as follows:
m6Am(Am)GGAGAAAAAUCACUCAGGGUCAAUGCCAGCGCUUCGUUAAUACAGAUGUAGGUGUUCCACAGGGUAGCCAGCAGCAUCCUGCGAUGCAGAUCCGGAACAUAAUGGUGCAGGGCGCUGACUUCCGCGUUUCCAGACUUUACGAAACACGGA.
The sequence of m6A probe was list as follows:
AUCUACCUGUCCAGUAGCCUUCAGGAUCAUGCUGUCUGACUUGCUGGm6ACAUCAUUCUAGUGCCAUAACUUCAGC.
Synthesize the dually modified m6Am&m6A probe
The dually modified m6Am & m6A probe was obtained by utilizing T4 RNA ligase 2 to ligate the 3′ end of RNA probe to the 5′ end of another RNA probe. The 30nt for m6Am probe was obtained by in vitro transcription reaction. For phosphorylation of 5′end of m6A probe, RNA was treated by PNK (NEB, M0201S) with incubation for 1 h at 37 °C. 1 μl of 20 μM m6Am probe, 2 μl of 20 μM m6A probe, 1.5 μl of 20 μM splint DNA (CTACTGGACAGGTAGATAGTCTTGAAGGATTC), 1 μl 10× T4 DNA ligase buffer and 3 μl nuclease-free water were heated at 65 °C for 3 min, followed by 5 min at 25 °C in a thermocycler. Add 1 μl of 10 units/μl T4 RNA ligase 2 (NEB, M0239S) and 0.5 μl RiboLock RNase inhibitor (ThermoFisher Scientific, EO0381) into the reaction mixture and incubate at 37 °C for 2 h. Synthetic m6Am & m6A probe was gel purified and sliced in the desired size. The sequence of the m6Am & m6A probe was listed as follows:
Capm6AmGGAUAGCAGGCAUGGAAUCCUUCAAGACUAUCUACCUGUCCAGUAGCCUUCAGGAUCAUGCUGUCUGACUUGCUGGm6ACAUCAUUCUAGUGCCAUAACUUCAGC.
In vitro probe assay
To investigate the FTO and ALKBH5 demethylation activity for m6Am and m6A, we performed in vitro demethylation assay using purified demethylases and synthesized probes contained m6A and m6Am. The demethylation assay was performed in 20 μl of mixture containing 200 ng probe, 50 mM MES (pH 6.5), 100 mM KCl, 2 mM MgCl2, 100 μM (NH4)2Fe(SO4)2·6H2O, 300 μM 2-ketoglutarate, 0 or 2 mM l-ascorbic acid, 0.4 U/ml SUPERase·In RNase inhibitor (Invitrogen, AM2694) and 2 μM purified FTO. The reaction mixture was incubated at 37 °C for 20 min. As the control, inactive demethylases were using for incubation with RNA.
CAGE-seq library preparation
CAGE allows mapping of the transcriptional start sites of capped RNAs. 5 μg of total RNA purified from HEK293T cells was used for the CAGE-seq45. Reverse transcription was carried out using PrimeScript reverse transcriptase (Takara, 2680A) with the RT-N15-EcoP primer. The RNA-cDNA hybrid was diol oxidized with NaIO4 and then the 5′ Cap and 3′ end of RNA were biotinylated. The noncapped biotinylated RNA-cDNA hybrid was eliminated by RNase digestion and the biotin-trapping step. The complete cDNA was released from RNA and then the Bar-coded 5′ linker was ligated to the single-stranded cDNA. The second-strand cDNA was synthesized using the biotin-modified primer. The cDNA was digested with EcoP15I (NEB, R0646S) and the 3′ linker was ligated to the 3′ end of cDNA. The cDNA was performed PCR amplification. The CAGE library was sequenced on Illumina HiSeq 2000. The sequences of primers and linkers were provided (Supplementary Data 6).
CAGE data analysis
CAGE tags were mapped to the human genome (hg19, UCSC Genome Browser) using TopHat2 (version 2.0.13) with default parameters that allow 0 mismatches per seed (22 nt), and CTSS pipeline was used to identify CAGE TSS45. Only uniquely mapped reads were used in downstream analysis within R and custom scripts. The mapped reads were sorted and the TSS tag was calculated. All unique 5′ ends of CAGE tag-supported TSS and the number of tags in each CTSS represents expression levels. Tag counts in each CTSS were normalized using tag pre million reads (TPMs). We also used Deeptools (version 3.5.0) to analyze the enrichment of m6Am in the −2 Kb to +2 Kb region around the CAGE TSS. The miCLIP-2015 m6Am list and miCLIP-2019 m6Am list were obtained from previous studies10,13.
Cloning, expression, and purification of FTO and ALKBH5
Wild-type human FTO gene and ALKBH5 gene with deletion of the 66 amino acids were cloned into pET-28a. The recombinant plasmids were then transduced into E. coli BL21 (DE3). FTO and ALKBH5 proteins were purified following the procedures as previously described46. Bacteria were grown at 37 °C until the OD600 reached 0.8–1, and then induced with 0.5 mM IPTG overnight at 15 °C. Cells were harvested by centrifugation and then resuspended in ice-cold lysis buffer (25 mM Tris-HCl pH 8.0, 150 mM NaCl, 10 mM imidazole), disrupted on ice, and centrifuged at 11,000×g for 1 h at 4 °C. The supernatant was purified on HiTrap His column (GE Healthcare) and eluted with buffer containing 25 mM Tris-HCl pH 8.0, 150 mM NaCl, 500 mM imidazole. Next, the fractions were purified by HiTrap Q column (GE Healthcare). Finally, the fractions were purified by Gel-filtration chromatography (HiLoad 16/600 Superdex 200 pg column, GE Healthcare) equilibrated in storage buffer containing 25 mM Tris-HCl pH 8.0, 150 mM NaCl, and 3 mM dithiothreitol (DTT). Purified protein was stored at −80 °C with the addition of 20% glycerol.
m6Am-seq
cap-m7G RNA immunoprecipitation
The total RNAs were extracted from control, heat shock-treated, and hypoxia-treated cells. 100 μg >200 nt RNA was fragmented into ~150 nt-long fragments using RNA fragmentation buffer (NEB, E6150S). Adding fragment stop solution buffer stopped the reaction, and fragments were purified and concentrated by ethanol precipitation. 10 ng of fragments were used as “input” and the remaining fragments were used for followed treatments. The fragmented RNA was denatured at 65 °C for 5 min, followed by chilling in ice. The denatured RNA was incubated overnight at 4 °C with 2 μg cap-m7G antibody (1:100 dilution) and 5 μl RiboLock RNase inhibitor in IPP buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% NP-40). 20 μl of protein A/G UltraLink Resin (ThermoFisher Scientific, 53132) were washed twice with IPP buffer, and then resuspended in a 200 μl IPP buffer contained 5 μl RiboLock RNase inhibitor. The antibody-RNA mixture was then incubated with protein A/G UltraLink Resin at 4 °C for 3 h. The Resin was washed twice with 1 ml of IPP buffer, once with 1 ml of low-salt IP buffer (10 mM Tris-HCl, pH 7.4, 75 mM NaCl, 0.05% NP-40), once with 1 ml of high-salt IP buffer (10 mM Tris-HCl, pH 7.4, 200 mM NaCl, 0.05% NP-40) and twice with 1 ml TET buffer (10 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.05% NP-40) at 4 °C. The beads-antibody-RNA was resuspended in 1 ml Trizol and rotated for 10 min at room temperature. cap-m7G-containing RNA fragments were extracted and precipitated. Around 150 ng cap-m7G-immunoprecipitated RNA fragments could be obtained.
In vitro demethylation treatment
1 ng of the cap-m7G-immunoprecipitated RNA was used as “m7G-IP”. RNA was subjected to [FTO (+)] or [FTO (−)] treatment. About 100 ng of RNA was used for [FTO (+)] treatment. RNA was denatured at 65 °C for 5 min, followed by chilling in ice. And then, the demethylation assay was performed in a 20 μl mixture which contained 50 mM MES (pH 6.5), 100 mM KCl, 2 mM MgCl2, 100 μM (NH4)2Fe(SO4)2·6H2O, 300 μM 2-ketoglutarate, 0.4 U/ml SUPERase·In RNase inhibitor and 1 μM purified FTO. After incubation at 37 °C for 20 min, the demethylated RNA was purified by phenol–chloroform extraction. The remaining RNA (around 50 ng) directly performed m6A RNA immunoprecipitation.
m6A RNA immunoprecipitation
The [FTO (−)] and [FTO (+)] samples were immunoprecipitated with m6A antibody. The m6A RNA immunoprecipitation was performed following the procedures as previously described21. 10 μl of protein A magnetic beads (ThermoFisher Scientific, 10002D) and 10 μl of protein G magnetic beads (ThermoFisher Scientific, 10004D) were mixed and washed twice with 500 μl of IPP buffer (10 mM Tris-HCl, pH 7.4,150 mM NaCl, 0.1% NP-40). The beads were resuspended in 500 μl of IPP buffer containing 2 μg of anti-m6A antibody (Millipore, ABE572). The mixture was incubated at 4 °C for at least 6 h. The beads-antibody mixture was washed twice in 500 μl of IPP buffer and resuspended with 200 μl mixture containing 40 μl of 5× IPP buffer, denatured RNA (the [FTO (−)] control and [FTO (+)] treated samples) and 5 μl of RNasin Plus RNase Inhibitor (Promega, N2615) at 4 °C for 2 h (1:100 dilution of anti-m6A antibody). The beads-antibody-RNA mixture was washed twice with 500 μl of IPP buffer, twice with 500 μl of low-salt IP buffer (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.1% NP-40), and twice with 500 μl of high-salt IP buffer (10 mM Tris-HCl, pH 7.5, 500 mM NaCl,0.1% NP-40). And then, bound fragments were eluted from beads with 200 μl of IPP buffer containing 6.7 mM N6-methyladenosine (Sigma-Aldrich, M2780) and 5 μl of RiboLock RNase inhibitor. An additional phenol–chloroform isolation and ethanol precipitation were used to purify the RNA. The bound RNA was called [FTO (+) m6A-IP] and [FTO (−) m6A-IP].
Library preparation and sequencing
The “input”, “m7G-IP”, [FTO (+) m6A-IP] and [FTO (−) m6A-IP] were subjected to library construction using SMARTer® Stranded Total RNA-Seq Kit v2—Pico Input Mammalian (634413, Takara – Clontech, Japan) according to the manufacturer’s protocol. The libraries were sequenced on Illumina HiSeq X10 with PE150.
Reads pre-processing and alignment
Strand orientation of the original RNA was preserved on the process of library construction and reads R2 yields sequences sense to the original RNA. Thus, only reads R2 was used in our study. Raw sequencing reads were firstly subjected to Trim_galore (version 0.6.6, http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and cutadapt software (version 1.18) for quality control and trimming adaptor. The quality threshold was set to 30, and the minimum length required for reads after trimming was 20 nt. Processed reads were mapped to genome (hg19, UCSC Genome Browser) using HISAT2 (version 2.1.0)47 with default parameters, separated by strand with in-house scripts, and reads coverage were showed by IGV. The mpileup files are generated from mapped BAM files using samtools mpileup command (version 1.9).
Transcriptome-wide identification of 5′-UTR peaks in m6A-IP sample
Mapped reads were subjected to the RNA methylation peak caller exomePeak48, the FOLD_ENRICHMENT was set to 3, the GENE_ANNO_GTF were set RefSeq_hg19 for peak calling. For genome-base peak caller MACS2 (version 2.1.1)49, the effective genome size was set to 2.7*109 for human, under the option of -nomodel and q-value cutoff 0.01. In order to identify high-confidence peak in m6A-IP sample, peak must meet the following conditions:(1) peak was overlapped between m6A-IP and m7G-IP samples, and (2) fold enrichment in m6A-IP higher than m7G-IP samples. Only peaks that can meet both requirements were used for m6A/m6Am signal identification in our study.
Definition and calculation of the demethylase-sensitivity peaks
In order to more accurately extract the m6Am peak and 5′-UTR m6A peak, we identify the demethylase-sensitivity peak between the FTO (+) and FTO (−) sample. Firstly, 5′-UTR peak defined in the previous section of “Transcriptome-wide identification of 5′-UTR peaks in m6A-IP sample” were used for analysis. Secondly, differential methylation peak between FTO (+) and FTO (−) samples were identified using exomePeak (P < 0.01) and DESeq2 (version 3.11)50 in the R environment (version 3.6). Thirdly, when the ratio of FTO (−) to FTO (+) is >2, is defined as demethylase-sensitivity peak.
Identification of m6Am and 5′-UTR m6A peaks
An m6Am peak was identified when: (1) peak was high-confidence in m6A-IP sample; (2) peak was demethylase sensitivity between the FTO (+) and FTO (−) sample. The 5′-UTR m6A peak was identified when: (1) peak was high-confidence in m6A-IP sample; (2) peak was demethylase insensitivity between the FTO (+) and FTO (−) sample, and was not decrease after FTO treatment.
Identification of m6Am site
To identify the potential m6Am site, we defined a “start rate difference” score (SRD score) for each nucleotide within an m6Am peak. SRD score takes into account normalized sequencing start rate in untreated samples m1 = [FTO (−) start reads]/[FTO (−) depth] and the reads coverage difference within the FTO (+) and FTO (−) samples m2 = [FTO (−) depth − FTO (+) depth]/[FTO (−) depth]. m1 reflects the fact that m6Am site is the first transcribed nucleotide, while m2 is a measure for demethylase sensitivity. Because the start rate in FTO (+) samples could be very low and hence prone to the high background, we used FTO (+) coverage in m2 instead of FTO (+) start rate.
The SRD score was used to identify the m6Am site. The defined a position i was defined to be m6Am when the following criteria were met: (1) position i must be located within m6Am peak and carry adenosine residue; (2) the start reads of position i are not <20 in FTO (−) sample; (3) the start rate of position i in FTO (−) sample is greater than that in m7G-IP sample; (4) the start reads coverage in m7G-IP sample is greater than that in input; (5) the position i possesses the top 3 of SRD scores (SRD score > 1).
Identification of stress-induced peak
In order to identify the stress-induced peak, differential methylation peaks between the untreated and stress sample were calculate using exomePeak (version 2.13, P < 0.01) and DESeq2. The stress-induced peak was defined, when the difference between the stress and untreated sample is >2.
Identification of PCIF1-dependent peak
The m6Am peak in WT and PCIF1-KO sample was calculated in section “Identification of m6Am and 5′UTR m6A peak”. To identification, the PCIF1-dependent peak, differential methylation peaks between WT and PCIF1-KO samples were calculate using exomePeak and DESeq2. The PCIF1-dependent peak was extracted, when the differential methylation ratio of WT to PCIF1-KO sample is >2 and P-value < 0.05.
Analysis of RNA-seq data
Paired-end, adapter-clean reads were aligned to the human genome (hg19, UCSC Genome Browser) using TopHat2 (version 2.0.13) with default parameters. The gene expression level was quantified as FPKM by Cufflinks (version 2.2.1)51.
Correlations analysis of m6Am site with histone modification and TSS
H3K27ac, H3K4me3, H3K4me1 and H3K9me3 ChIP-seq data was downloaded from ENCODE portal52 (https://www.encodeproject.org/) with the following identifiers: ENCSR000DTU, ENCSR000FCJ, ENCSR000FCH, ENCSR000FCG. GRO-seq data were downloaded from the GEO database (GSE92375)53. We divided the m6Am sites into two parts with and without BCA motif, m6Am sites was extended 500 bp for each site, the overlapped site was removed. Deeptools were used to analyze ChIP-seq enrichment in −5 Kb to +5 Kb region around the m6Am sites.
Motif discovery and GO enrichment analysis
Peaks were annotated by homer software (version 4.10) with hg19 reference genome to annotate the marked genes. For the analysis of sequence consensus, the top 1000 peaks were chosen for de novo motif analysis with MEME (version 4.12.0)54, with 100 nt-long peak summit centered sequences as input. Gene Ontology (GO) enrichment analyses were performed using DAVID web-based tool (version 6.8)55.
Statistical analysis and quantification
For the statistical analysis of results, statistical evaluation was performed by unpaired two-sided Mann–Whitney U-test.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The sequence data generated in this study have been deposited in the NCBI GEO, under accession code GSE180253 that is publicly accessible at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE180253. For H3K27ac, H3K4me3, H3K4me1, and H3K9me3 ChIP-seq public data were downloaded from ENCODE portal52 (https://www.encodeproject.org/) with the following identifiers: ENCSR000DTU, ENCSR000FCJ, ENCSR000FCH, and ENCSR000FCG. GRO-seq data were downloaded from the GEO database (GSE92375)53. Source data are provided with this paper.
Code availability
Custom code used for m6Am-seq analysis is available on GitHub (https://github.com/KaiLi-bioinfo/m6Am_seq/).
References
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307 (2018).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Frye, M., Jaffrey, S. R., Pan, T., Rechavi, G. & Suzuki, T. RNA modifications: what have we learned and where are we headed? Nat. Rev. Genet. 17, 365–372 (2016).
Gilbert, W. V., Bell, T. A. & Schaening, C. Messenger RNA modifications: form, distribution, and function. Science 352, 1408–1412 (2016).
Wei, C., Gershowitz, A. & Moss, B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature 257, 251–253 (1975).
Keith, J. M., Ensinger, M. J. & Moss, B. HeLa cell RNA (2′-O-methyladenosine-N6-)-methyltransferase specific for the capped 5′-end of messenger RNA. J. Biol. Chem. 253, 5033–5039 (1978).
Akichika, S. et al. Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363, eaav0080 (2019).
Sun, H., Zhang, M., Li, K., Bai, D. & Yi, C. Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 29, 80–82 (2019).
Sendinc, E. et al. PCIF1 catalyzes m(6)Am mRNA methylation to regulate gene expression. Mol. Cell 75, 620–630.e629 (2019).
Boulias, K. et al. Identification of the m(6)Am methyltransferase PCIF1 reveals the location and functions of m(6)Am in the transcriptome. Mol. Cell 75, 631–643 e638 (2019).
Fan, H. et al. PCIF1, a novel human WW domain-containing protein, interacts with the phosphorylated RNA polymerase II. Biochem. Biophys. Res. Commun. 301, 378–385 (2003).
Mauer, J. et al. Reversible methylation of m(6)Am in the 5′ cap controls mRNA stability. Nature 541, 371–375 (2017).
Wei, J. et al. Differential m(6)A, m(6)Am, and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e975 (2018).
Li, X., Xiong, X. & Yi, C. Epitranscriptome sequencing technologies: decoding RNA modifications. Nat. Methods 14, 23–31 (2016).
Dominissini, D. et al. Topology of the human and mouse m(6)A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K. D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Zhou, J. et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594 (2015).
Meyer, K. D. et al. 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010 (2015).
Coots, R. A. et al. m(6)A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514 e507 (2017).
Linder, B. et al. Single-nucleotide-resolution mapping of m(6)A and m(6)Am throughout the transcriptome. Nat. Methods 12, 767–772 (2015).
Liu, J. et al. Landscape and regulation of m(6)A and m(6)Am methylome across human and mouse tissues. Mol. Cell 77, 426–440 e426 (2020).
Jia, G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887 (2011).
Zheng, G. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29 (2013).
Young, J. I., Zuchner, S. & Wang, G. Regulation of the epigenome by vitamin C. Annu. Rev. Nutr. 35, 545–564 (2015).
Li, X. et al. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat. Chem. Biol. 11, 592–597 (2015).
Li, X. et al. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat. Chem. Biol. 12, 311–316 (2016).
Chan, C. T. et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247 (2010).
Lee, P., Chandel, N. S. & Simon, M. C. Cellular adaptation to hypoxia through hypoxia inducible factors and beyond. Nat. Rev. Mol. Cell Biol. 21, 268–283 (2020).
Blais, J. D. et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol. Cell. Biol. 24, 7469–7482 (2004).
Bi, M. et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 24, 3470–3481 (2005).
Semenza, G. L. Hypoxia-inducible factors in physiology and medicine. Cell 148, 399–408 (2012).
An, W. G. et al. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature 392, 405–408 (1998).
Zhao, L. Y., Song, J., Liu, Y., Song, C. X. & Yi, C. Mapping the epigenetic modifications of DNA and RNA. Protein Cell 11, 792–808 (2020).
Yu, M. et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149, 1368–1380 (2012).
Liu, Y. et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat. Biotechnol. 37, 424–429 (2019).
Zheng, G. et al. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods 12, 835–837 (2015).
Cozen, A. E. et al. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods 12, 879–884 (2015).
Li, X. et al. Base-resolution mapping reveals distinct m(1)A methylome in nuclear- and mitochondrial-encoded transcripts. Mol. Cell 68, 993–1005.e1009 (2017).
Blaschke, K. et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222–226 (2013).
Incarnato, D. et al. High-throughput single-base resolution mapping of RNA 2′-O-methylated residues. Nucleic Acids Res. 45, 1433–1441 (2017).
Dai, Q. et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods 14, 695–698 (2017).
Bird, J. G. et al. The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016).
Karijolich, J. & Yu, Y. T. Spliceosomal snRNA modifications and their function. RNA Biol. 7, 192–204 (2010).
Huang, H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295 (2018).
Takahashi, H., Lassmann, T., Murata, M. & Carninci, P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 7, 542–561 (2012).
Jia, G. et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582, 3313–3319 (2008).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Meng, J. et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods 69, 274–281 (2014).
Feng, J. X., Liu, T., Qin, B., Zhang, Y. & Liu, X. S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7, 1728–1740 (2012).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Sloan, C. A. et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 44, D726–D732 (2016).
Bouvy-Liivrand, M. et al. Analysis of primary microRNA loci from nascent transcriptomes reveals regulatory domains governed by chromatin architecture. Nucleic Acids Res. 45, 12054 (2017).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Acknowledgements
We would like to thank Prof. Guifang Jia and Mr. Wei Zhang for their assistance with recombinant FTO and ALKBH5 plasmids, and Xiaoyu Li and Xushen Xiong for discussions. We thank the National Center for Protein Sciences at Peking University in Beijing, China for assistance with RNA quality control and High Performance Computing Platform of the Center for Life Science for assistance with the analysis. This work was supported by the National Key R&D Program of China (Nos. 2019YFA0110900 and 2019YFA0802200), the National Natural Science Foundation of China (Nos. 91940304, 21825701, 31861143026, and 91740112 to C.Y.).
Author information
Authors and Affiliations
Contributions
H.S. and C.Y. conceived the project, designed the experiments; H.S., K.L., and C.Y. wrote the manuscript; H.S. performed the experiments with the help of M.Z. and J.L.; K.L. designed and performed the bioinformatics analysis with the help of X.Z. and H.M.; J.L. performed the CAGE experiment; M.Z. performed the PCIF1-KO cell line and participated in the discussion.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sun, H., Li, K., Zhang, X. et al. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nat Commun 12, 4778 (2021). https://doi.org/10.1038/s41467-021-25105-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-021-25105-5
This article is cited by
-
EMDL_m6Am: identifying N6,2′-O-dimethyladenosine sites based on stacking ensemble deep learning
BMC Bioinformatics (2023)
-
RNA methylations in hepatic fibrosis, a gradually emerging new treatment strategy
Cell & Bioscience (2023)
-
PCIF1, the only methyltransferase of N6,2-O-dimethyladenosine
Cancer Cell International (2023)
-
Regulation and functions of non-m6A mRNA modifications
Nature Reviews Molecular Cell Biology (2023)
-
Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI
Nature Biotechnology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.