Abstract
Anthocyanin biosynthesis and sugar metabolism are important processes during plant growth, but the molecular interactions underlying these pathways are still unclear. In this work, we analyzed the anthocyanin and soluble sugar contents, as well as the transcript levels of transcription factors that are known to be related to the biosynthesis of anthocyanin in ‘Hongcui 1’ apple flesh during fruit development. Overexpression of MdMYB6 in red-fleshed calli was found to reduce anthocyanin content and result in downregulated expression of the MdANS and MdGSTF12 proteins. Yeast one-hybrid and electrophoretic mobility shift analyses showed that MdMYB6 could directly bind to the promoters of MdANS and MdGSTF12, indicating that MdMYB6 could inhibit anthocyanin biosynthesis by regulating MdANS and MdGSTF12. Overexpression of MdTMT1 in the Arabidopsis tmt1 mutant restored the glucose and fructose contents to the wild-type levels, while overexpression of MdTMT1 in red-fleshed calli increased the contents of glucose and fructose but reduced the contents of UDP-glucose, UDP-galactose, and anthocyanin. Using a GUS reporter system, yeast one-hybrid, chromatin immunoprecipitation-PCR and electrophoretic mobility shift analyses, we found that MdMYB6 could bind to the promoter of MdTMT1, resulting in increased promoter activity. Overexpression of MdMYB6 in calli overexpressing MdTMT1 increased the expression of MdTMT1, which led to reduced contents of UDP-glucose and UDP-galactose and decreased anthocyanin content compared to those of the calli that overexpressed MdTMT1. This finding suggested that MdMYB6 could also inhibit anthocyanin biosynthesis by regulating MdTMT1 to decrease the contents of UDP-glucose and UDP-galactose. Taken together, these results showed that MdMYB6 and MdTMT1 play key roles in both anthocyanin biosynthesis and sugar transport.
Similar content being viewed by others
Introduction
Anthocyanins are important secondary metabolites in plants and are mainly distributed in the vacuoles of plant tissues, including seeds, fruits, flowers, leaves and other organs1. Anthocyanins accumulate at high levels in some mature fruits, such as grapes, strawberries, apples, red pears, mangosteen and blood oranges. These pigments not only give fruits their rich colors but have also been shown to prevent cardiovascular disease, reduce obesity and improve glucose homeostasis in humans2,3. Anthocyanin biosynthesis involves various transcription factors (TFs) and structural enzymes, which are encoded by genes such as anthocyanin synthase (ANS) and UDP-glucose flavonoid 3-glu-cosyltransferase (UFGT)4,5. This process is transcriptionally regulated by the MYB-bHLH-WD40 complex, whose members have been studied extensively6,7,8,9. Homologs of Arabidopsis thaliana AtMYB75 that are involved in anthocyanin biosynthesis have been found in pear (PyMYB10), peach (PpMYB10), grape (VvMYBA1 and VvMYBA2), strawberry (FaMYB10), and apple (MdMYB1)10,11,12. In red-fleshed and red-foliage apple, the transcription factor encoded by MdMYB10 can bind to its own promoter and control anthocyanin biosynthesis in apple flesh13,14. A novel MYB gene, MdMYB110a, was found to regulate anthocyanin biosynthesis in sangrado apple that has green foliage and develops red flesh in the fruit cortex late in maturity15. In addition, the first MYB repressor involved in anthocyanin biosynthesis is FaMYB1 in strawberry, which reduces anthocyanin content in tobacco petals when heterologously expressed in tobacco16. A petunia homologue, PhMYB27, was shown to act upon MBW complexes to inhibit anthocyanin biosynthesis17. In apple, MdMYB6 could inhibit the anthocyanin content in transgenic Arabidopsis18, and MdMYB16 could directly repress anthocyanin biosynthesis via the C-terminal EAR sequence19.
Anthocyanin biosynthesis is induced by plant hormones and some environmental factors, such as low temperature and ultraviolet-B radiation20,21. In apples, MdbHLH3 has been shown to be involved in low temperature-induced anthocyanin biosynthesis22, while MdCOL11 and MdBBX20 are known to control anthocyanin biosynthesis in response to ultraviolet B and low temperature23,24. MdMYB9 and MdMYB11 participate in jasmonic acid-induced anthocyanin biosynthesis25. In addition, AtTT19 and MdGSTF6 transport anthocyanins from the cytosol to the vacuole, thereby contributing to anthocyanin accumulation in plant cells26,27.
Sugars are the main products of photosynthesis in plants and are involved in nutrient storage and transport, signal transduction, and stress resistance28. In members of Rosaceae, such as apple, apricot, peach and pear, sorbitol is the main form of sugar subjected to long-distance transport via the phloem. However, fruits contain starch, sucrose, fructose, and glucose as the main sugars, and sorbitol only makes up a small portion of all the sugars. In Arabidopsis, there are at least 69 sugar transporters, including monosaccharide and polysaccharide transporters, belonging to eight subfamilies29. Sucrose transporters comprise five subfamilies, namely, SUT1–530,31. SUT1 and SUT3 are mainly expressed in both source and sink tissues and are responsible for the loading and unloading of sucrose in the phloem and the absorption of sucrose by sink tissues32. Endler et al.33 showed that HvSUT2 and AtSUT4 are localized at the tonoplast and mediate the exchange of sucrose between the vacuole and the cytoplasm.
Plant monosaccharide transporters are membrane-bound proteins with 12 transmembrane domains34, which act to transport glucose, fructose, galactose, mannose and inositol across cell membranes and tonoplasts. In Arabidopsis, the monosaccharide/proton symporter AtSTP1 transports glucose and is expressed in germinated seeds, roots, and cotyledon guard cells, where it participates in sugar absorption under dark conditions in many tissues and in roots under normal physiological conditions35. The early response to dehydrate 6-like protein (AtERD6L) is localized to the tonoplast and transports glucose stored in the vacuole to the cytoplasm under stress conditions36. The tonoplast monosaccharide transporters (TMT) AtTMT1 and AtTMT2 were found to be induced by exogenous sugar treatment and are involved in vacuolar monosaccharide transport, especially during responses to salt and cold stresses34,37. Members of the TMT subfamily have phosphorylation sites in the central loop between the sixth and seventh transmembrane domains, which can be phosphorylated under low temperature to increase the transport of glucose and fructose to the vacuole38.
Previous studies have demonstrated that sugars increase the expression levels of genes associated with anthocyanin biosynthesis in Petunia hybrid39 and induce F3H expression to increase anthocyanin content in grape berries40 In Arabidopsis, the sucrose transporter AtSUC1 plays a key role in sucrose-induced anthocyanin accumulation41, while in apple, the biosynthetic pathway for UDP sugar contributes to the biosynthesis of cyanidin 3-galactoside in fruit skin42. However, the molecular mechanisms underlying the relationship between sugar biosynthetic pathways and anthocyanin accumulation in apple are still unclear.
In this work, we found a monosaccharide transporter, MdTMT1, that localizes to the tonoplast. Overexpression of MdTMT1 in the Arabidopsis tmt1 mutant or red-fleshed apple callus increased the contents of glucose and fructose, while MdTMT1 inhibited anthocyanin biosynthesis by decreasing the amounts of UDP-galactose (UDP-gal) and UDP-glucose (UDP-glu). The results of GUS analyses, yeast one-hybrid, chromatin immunoprecipitation-PCR (ChIP-PCR), and electrophoretic mobility shift analyses (EMSA) revealed that MdMYB6 can directly bind to the promoter region of MdTMT1 and lead to upregulation of gene expression. Overexpression of MdMYB6 in the calli overexpressing MdTMT1 increased the expression of MdTMT1, which led to reduced UDP-glu and UDP-gal contents. In addition, MdMYB6 was shown to bind the promoters of MdGSTF12 and MdANS and inhibit their expression. In summary, MdMYB6 suppresses anthocyanin biosynthesis both by directly inhibiting MdGSTF12 and MdANS and by reducing UDP-glu and UDP-gal contents by regulating MdTMT1.
Results
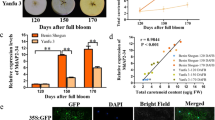
Contents of soluble sugars and anthocyanin and the expression levels of transcription factors associated with anthocyanin biosynthesis in ‘Hongcui 1’ apple during fruit development
To examine the anthocyanin levels, sugar contents and transcription factor expression levels during ‘Hongcui 1’ apple maturation, we took cross sections of the apple flesh throughout development. Overall, the flesh color became lighter and the anthocyanin content gradually decreased over the course of development (Fig. 1a, b), while the transcript levels of genes encoding regulatory factors that positively affect anthocyanin biosynthesis, such as MdMYB9, MdMYB10, MdMYB11, MdMYBPA1, MdMYB110a, MdbHLH3 and MdbHLH33, were also reduced during fruit development. Moreover, the transcript levels of genes encoding negative regulatory factors, including MdMYB6 and MdMYB16, increased during fruit development (Fig. 1c). The contents of soluble sugars in the flesh of ‘Hongcui 1’ apple, including glucose, fructose and sucrose, increased during development (Fig. 1b). Although a previous study detected a positive correlation between the coloration of ‘Fuji’ apple skin and the concentration of soluble sugars in apple flesh43, we considered it unintuitive to compare apple skin with apple flesh. Our results indicated that there might be a negative correlation between anthocyanin content and soluble sugar content in ‘Hongcui 1’ apple flesh.
Quantitative proteomic comparison between calli expressing green fluorescent protein (GFP) and calli expressing MdMYB6-GFP
To investigate the function of MdMYB6 in regulating sugar and anthocyanin levels, we generated MdMYB6 overexpression lines. In the red-fleshed calli, the overexpression of MdMYB6 led to a change in color from red to pink, indicating reduced anthocyanin content (Fig. 2a,b). To identify differentially expressed proteins involved in this color change, we compared the proteomes of the calli overexpressing GFP (control) and the calli overexpressing MdMYB6-GFP using the tandem mass tags (TMT) quantitative proteomics technique (Zhongke New Life Co., Shanghai, China). In total, 4865 proteins were identified in the self-built library. According to the criteria for differential expression (>1.5-fold differences in expression levels between the two libraries, and P values < 0.05), we found 609 upregulated proteins and 477 downregulated proteins in the control versus the MdMYB6-GFP overexpression line (Fig. 2c).
a Red-fleshed callus and callus overexpressing MdMYB6. b Transcript levels of MdMYB6 and anthocyanin content. c Volcano plot of all differentially expressed proteins. The x-axis represents significant differences between A and B after log2 transformation. The y-axis represents significant differences in the P value after log10 transformation. Red dots represent upregulated proteins, while blue dots represent downregulated proteins in A vs. B. d KEGG pathway enrichment analysis of the proteins that were differentially expressed between A and B. The y-axis represents significantly enriched KEGG pathways, while the x-axis indicates the number of differentially expressed proteins in the KEGG pathways. The color gradient represents the P value, where red denotes small p values and more significant KEGG pathway enrichment. e Enriched GO terms of proteins that were differentially expressed between A and B. The x-axis represents the enriched GO functional classification, which is divided into three categories: biological process, molecular function and cellular component, while the y-axis indicates the number of differentially expressed proteins under each functional classification. The color gradient represents the P value, where red denotes small P values and more significant GO functional classification enrichment.
The GO functional analysis of the proteins that were differentially expressed between the control and the MdMYB6-GFP overexpression line found that several important biological terms, such as biological process, molecular function and cellular component, were enriched (Fig. 2e). The results of KEGG pathway analysis showed that significant differences had occurred in the ribosome, photosynthesis, pancreatic secretion and other pathways (Fig. 2d). In the calli overexpressing MdMYB6-GFP, the upregulated proteins included MdMYB6, MdTMT1, MdWRKY33 and MdVPS11, and the downregulated proteins included MdANS, MdFLS, MdGSTF12, MdCHS, and MdF3H (Fig. 3a, b).
a Heatmap-cluster analysis of the 23 proteins that were differentially expressed between A and B. Boxes denote protein expression levels after log2 conversion, while colors indicate the Z-score values of normalized protein expression. b Descriptions of the 23 screened proteins and their relative protein expression levels in A and B.
Interactions between MdMYB6 and the promoters of MdANS and MdGSTF12
The results of the proteomic analysis indicated that MdANS and MdGSTF12, which are key enzymes involved in anthocyanin biosynthesis, were downregulated in the calli overexpressing MdMYB6-GFP5,27. Therefore, we isolated the 1692-bp promoter of MdANS and the 1710-bp MdGSTF12 promoter sequence to identify target DNA binding site(s) of MdMYB6 using yeast one-hybrid (Y1H) assays (Fig. 4a). Recombinant pHIS2 plasmids containing the MdANS and MdGSTF12 promoters were transformed into Y187 yeast. Each strain was then cultivated in –tryptophan–histidine (–T–H) selective media containing a series of different 3-amino-1,2,4-triazole (3-AT) concentrations, which resulted in a reduction of yeast growth as the 3-AT concentration increased.
a The 1692-bp sequence of the MdANS promoter and the 1710-bp sequence of the MdGSTF12 promoter. b Screening of different concentrations of 3-AT to determine the optimum reporter gene inhibition. c Yeast one-hybrid analysis of the interactions between the MdMYB6 protein and the promoters of MdANS and MdGSTF12. d Electrophoretic mobility shift analysis of the interactions between the MdMYB6 protein and the promoters of MdANS and MdGSTF12. Lane 1 includes the labeled DNA probes with no protein added, while lane 2 includes protein as well as the labeled DNA probes without any competitor. Unlabeled DNA probes were added in increasing amounts (25×, 50×, and 100×) in lanes 3, 4, and 5. Unlabeled mutant DNA probes (lanes 7, 8, and 9) were added as competitors. In lane 6, labeled mutant DNA probes and protein were added but with no competitor.
The yeast strain containing the pHIS2-MdANSpro plasmid was not able to grow when using –T–H selective medium containing 80 mM 3-AT, while the yeast strain with the pHIS2-MdGSTF12pro plasmid could not grow on the –T–H selective medium with 100 mM 3-AT (Fig. 4b). Thus, 80 mM 3-AT could reduce the expression of the HIS3 reporter when its expression was controlled by the MdANS promoter, and 100 mM 3-AT could inhibit the expression of the HIS3 reporter gene when it was placed under the control of the MdGSTF12 promoter. Next, various combinations of pGADT7-MdMYB6 with pHIS2-MdANSpro and pHIS2-MdGSTF12pro were cotransformed into Y187 yeast. The yeast strain containing both pGADT7-MdMYB6 and pHIS2-MdANSpro grew on -tryptophan–histidine–leucine (–T–H–L) selective media supplemented with 80 mM 3-AT, while the yeast strain containing both pGADT7-MdMYB6 and pHIS2-MdGSTF12pro grew on –T–H–L selective media that contained 100 mM 3-AT. The yeast strains containing combinations of pGADT7 plasmids driven by pHIS2-MdANSpro and pHIS2-MdGSTF12pro were unable to grow on any of these selective media (Fig. 4c).
Using tools in the PlantCARE database, we found one MYB recognition element (MRE) that was present in the promoter of MdANS and two MYB binding site (MBS) elements in the MdGSTF12 promoter. Therefore, we used electrophoretic mobility shift assays (EMSAs) to analyze the interaction between the MdMYB6 protein and these elements. In the EMSAs, the mixture of MdMYB6 protein and biotin-labeled probes containing the identified MRE yielded a dense band, which disappeared with increasing concentrations of the probe without the biotin label. The mixture of MdMYB6 protein and a biotin-labeled probe containing a mutated MRE did not yield a band in this analysis (Fig. 4d). Similar results were observed in an EMSA with a mixture of MdMYB6 protein and a biotin-labeled probe containing the MBS element (Fig. 4d). Taken together, the results of the EMSAs showed that MdMYB6 can bind to the promoters of both MdANS and MdGSTF12.
Protein sequence analysis, phylogenetic tree construction, and subcellular localization of MdTMT1
Apple tonoplast monosaccharide transporter 1 (TMT1), which was differentially expressed between the calli overexpressing GFP and the calli overexpressing MdMYB6-GFP, was subjected to KEGG pathway and functional annotation analyses. MdTMT1 has 739 amino acid residues and shows 63.88% similarity to AtTMT1, 71.18% similarity to OsTMT1, and 80.32% similarity to VvTMT1 at the amino acid sequence level (Fig. 5a). We also detected strong similarities of the transmembrane domains among these four TMT proteins. The MdTMT1 protein was predicted to contain 12 transmembrane domains and a large central hydrophilic loop that spans the region that connects transmembrane domains 6 to 7 (Fig. 5a). This domain architecture was highly similar to the structures of AtTMT1, OsTMT1 and VvTMT134.
a Comparison of the predicted protein structures of MdTMT1, AtTMT1, CsTMT1, and VvTMT1. b Phylogenetic tree of proteins related to sugar transport. (c) Subcellular localization analysis of MdMYB6-GFP in apple callus protoplasts. The first panel shows a GFP fluorescence image, the second panel shows a bright field image, and the last panel shows the merged GFP fluorescence and bright field image. d Subcellular localization analysis of GFP in apple callus protoplasts. The details of the 3 panels are the same as in c.
Next, we constructed a phylogenetic tree based on the protein sequences of MdTMT1 and other sugar transporters. This tree resulted in four groups: members of the STP family, members of the TMT family (including MdTMT1), members of the SWEET family, and members of the SUT/SUC family (Fig. 5b).
In the subcellular localization analyses, the fluorescence of GFP fused with MdTMT1 was only observed in the tonoplast of protoplasts (Fig. 5c), while that of the unfused GFP was detected throughout the whole protoplast (Fig. 5d). These results clearly indicate that MdTMT1 is localized to the tonoplast.
MdTMT1 overexpression increased the glucose and fructose contents in Arabidopsis and the red-fleshed apple calli
To test the function of MdTMT1, we determined the contents of sucrose, glucose, and fructose in 40-day-old plants of the wild-type Arabidopsis (WT), the TMT1 mutant of Arabidopsis (tmt1), and tmt1 overexpressing MdTMT1 (tmt1 + MdTMT1) (Fig. 6a). Although there was no difference in the sucrose content among these three lines, the contents of glucose and fructose were lower in the tmt1 plants than in the WT plants. Overexpression of MdTMT1 in tmt1 restored the contents of glucose and fructose to some extent (Fig. 6b). Overexpressing MdTMT1 in red-fleshed calli increased the contents of glucose and fructose but not that of sucrose. When MdTMT1 was silenced in the red-fleshed calli using RNAi, the contents of glucose and fructose were reduced (Fig. 7a, c). Finally, overexpressing MdMYB6 in the MdTMT1-overexpressing calli increased the transcript level of MdTMT1 (Fig. 7a, b).
Significance tests are shown as i, ii, iii, and iv. Different letters above the columns denote significant differences (P < 0.01). a Red-fleshed apple callus (Rc), red-fleshed apple callus overexpressing MdTMT1 (OETMT1), red-fleshed apple callus with silenced MdTMT1 (RNAi: TMT1) and red-fleshed apple callus co-overexpressing MdMYB6 and MdTMT1 (OE(TMT1 + MYB6)). b Transcript levels of MdMYB6 and MdTMT1 in the above four calli. Endogenous MdTMT1 (endoTMT1), exogenous MdTMT1 (exoTMT1). c Contents of sucrose, glucose, and fructose in the four calli.
MdMYB6 bound to and activated the MdTMT1 promoter
Next, we utilized the 1520-bp sequence of the MdTMT1 promoter for the GUS reporter assay and Y1H analysis (Fig. 8a). In the GUS histochemical staining analyses, the callus with both the MdTMT1 promoter and MdMYB6 was more deeply stained than the callus with only the MdTMT1 promoter (Fig. 8b). Consistent with this result, GUS activity was higher in the calli with both the MdTMT1 promoter and MdMYB6 than in the calli with only the MdTMT1 promoter (Fig. 8c). In the Y1H assay, MdMYB6 could bind to the promoter of MdTMT1 (Fig. 8d).
a The 1520-bp sequence of the MdTMT1 promoter. b GUS histochemical staining of a callus harboring a plasmid containing the MdTMT1 promoter and GUS reporter gene (TMT1 pro: GUS) and a callus overexpressing MdMYB6 harboring the TMT1 pro:GUS vector (TMT1 pro: GUS + MYB6). c Analysis of GUS activity. d Yeast one-hybrid analysis of the interaction between the MdYB6 protein and the MdTMT1 promoter. e Predicted elements bound by MdMYB6 in the MdTMT1 promoter. f ChIP-PCR analyses of the interaction between the MdTMT1 promoter fragments and the MdMYB6 protein. MdMYB6-DNA complexes were then coimmunoprecipitated from the calli overexpressing MdMYB6-GFP via a GFP antibody. g EMSA analysis of the interaction between the MdTMT1 promoter and MdMYB6 protein. Lane 1 included labeled DNA probes with no added protein, while lane 2 included labeled DNA probes with no competitor or protein. Unlabeled DNA probes were added in increasing amounts (25×, 50×, and 100×) in lanes 3, 4, and 5. Unlabeled mutant DNA probes (lanes 7, 8, and 9) were added as competitors. Lane 6 included labeled mutant DNA probes and protein but without competitor.
Using tools from the PlantCARE database, we found six MYBCORE and one MBS elements within the 1520-bp sequence of the MdTMT1 promoter (Fig. 8e). The sequences from −492 to −688, −700 to −874, and −908 to −1084 were termed TMT1 pro-1, TMT1 pro-2, and TMT1 pro-3, respectively. TMT1 pro-1 contained one MYBCORE element, TMT1 pro-2 contained two MYBCORE elements, and TMT1 pro-3 contained one MBS and three MYBCORE elements. These three promoter regions were amplified for ChIP-PCR analyses. As expected, the input of GFP and MdMYB6-GFP all produced bands in analyses for the three promoter regions, the IP of GFP alone produced no band and that of MdMYB6-GFP produced a band corresponding to TMT1 pro-2 (Fig. 3b). These results indicate that MdMYB6 can bind to the sequence from −700 to −874 in the MdTMT1 promoter (Fig. 8f). In the EMSAs, MdMYB6 could bind to only one MYBCORE element (CCGTTA) in the sequence from −830 to −835 (Fig. 8g). Taken together, these analyses showed that MdMYB6 binds to the MYBCORE element in the promoter of MdTMT1 and increases its activity.
MdMYB6 and MdTMT1 may be involved in anthocyanin metabolism
TMT proteins are transmembrane sugar transporters34,37, but overexpression of MdTMT1 in red-fleshed apple callus changed the color from red to light red, indicating that MdTMT1 can also affect anthocyanin content (Fig. 9a, b). In our analyses, MdMYB6 inhibited anthocyanin biosynthesis (Fig. 2a, b) and affected the expression of MdTMT1 (Figs. 3a, b and 7b). To rule out the possibility of an inhibitory effect of endogenous MdMYB6, we used RNA interference (RNAi) technology to silence MdMYB6 expression.
Significance tests are labeled i, ii, iii, and iv, with different letters above columns denoting significant differences (P < 0.01). a Red-fleshed apple callus (Rc), red-fleshed apple callus overexpressing MdTMT1 (OETMT1), red-fleshed apple callus with silenced MdMYB6 and overexpression of MdTMT1 (RNAi: MYB6 + OETMT1), red-fleshed apple callus co-overexpressing MdMYB6 and MdTMT1 (OE(TMT1 + MYB6)). b Anthocyanin levels in all four calli. c Transcript levels of key genes in the four transfected calli. d Contents of UDP-glu and UDP-gal in the four calli.
Overexpression of MdTMT1 in the calli with silenced MdMYB6 still led to a change in the color of the calli from red to light red (Fig. 9a), which was associated with reduced anthocyanin levels (Fig. 9b). This finding indicates that MdTMT1 functioned independently in inhibiting anthocyanin accumulation. Overexpression of MdMYB6 in the red-fleshed calli led to reduced expression of MdANS and MdGSTF12 and increased expression of MdTMT1 (Fig. 9c). Overexpression of MdTMT1 in the red-fleshed callus decreased the expression of MdUGPase (Fig. 9c) and the contents of UDP-glu and UDP-gal (Fig. 9d), while overexpression of MdMYB6 in the callus overexpressing MdTMT1 decreased the contents of UDP-glu and UDP-gal (Fig. 9d).
Discussion
MdMYB6 specifically binds to the promoters of MdANS and MdGSTF12 and inhibits anthocyanin biosynthesis
MYB transcription factors (TFs) play critical roles in anthocyanin metabolism by controlling the expression levels of several genes encoding key structural enzymes involved in anthocyanin biosynthesis44. Previous studies have shown that the promoter of MdANS can be stimulated by the binding of both MdMYB9 and MdMYB1125. Several MYB proteins have been shown to recognize and bind to MYBCORE, MRE, and MBS elements in gene promoters45,46,47. In apple, we found that MdMYBPA1 binds to the MRE in the promoter of MdANS and to the MBS element in the promoter of MdUFGT, resulting in increased transcription and higher anthocyanin content47.
There are two types of MYB TFs that have been shown to reduce anthocyanin biosynthesis, one of which is dependent on its own EAR inhibitory sequence, while the other is independent19,48,49,50. In addition, there are two mechanisms by which MYB repressors control anthocyanin biosynthesis: they can act upon MBW complexes (FaMYB151 and MdMYB15L52) or directly bind to the promoters of target genes (AtMYB453 and MdMYB1619).
In our study, overexpression of MdMYB6 in the callus of red-fleshed apple decreased the anthocyanin content (Fig. 2b) and downregulated MdANS (Figs. 3a, b and 9c). The Y1H assay revealed that MdMYB6 can bind to the MdANS promoter (Fig. 4a, b, c), while the EMSA analysis showed that MdMYB6 can recognize and bind to the MRE in the promoter of MdANS (Fig. 4d). This finding was consistent with a previous study that showed that MdMYB6 could suppress the expression of AtANS in transgenic Arabidopsis18. Recent studies have shown that anthocyanins are synthesized in the cytoplasm and need to be moved to the vacuole for storage. Glutathione S-transferase (GST) was reported to participate in the transport of anthocyanin, such as transparent testa 19 (AtTT19) in Arabidopsis26, LcGST4 in litchi54 and MdGSTF6 in apple27. In this study, the comparative proteomic analysis between the calli overexpressing GFP and those overexpressing MdMYB6-GFP revealed that the protein level of MdGSTF12 was downregulated in the calli overexpressing MdMYB6-GFP (Fig. 3). Overexpression of MdMYB6 in the red-fleshed callus reduced the transcript level of MdGSTF12 (Fig. 9c), while the Y1H and EMSA analyses demonstrated that MdMYB6 can directly bind to the MBS element in the MdGSTF12 promoter (Fig. 4). Together, these results suggest that MdMYB6 inhibits anthocyanin biosynthesis by binding to the promoters of both MdANS and MdGSTF12, resulting in reduced transcriptional activity.
MdMYB6 promotes the expression of MdTMT1, which transports glucose and fructose across the tonoplast
The vacuole is the largest subcellular organelle in plant cells, accounting for more than 90% of the total cell volume. It is the storage site for most metabolites and soluble sugars in plant cells36,55. Recent studies on sugar transporters have revealed some of the mechanisms of sugar transport across the tonoplast. For example, tonoplast-localized TMT-like monosaccharide transporters mediate the transport of monosaccharides into the vacuole via a reverse transport mechanism coupled with protons34,37,56. The TMT proteins generally have twelve transmembrane domains and a long loop connecting the sixth and seventh transmembrane domains38. In Arabidopsis, a subcellular localization assay showed that AtTMT1 is localized at the tonoplast. The contents of glucose and fructose in leaves were found to be significantly lower in the AtTMT1 mutant plants than in the wild-type plants, indicating that AtTMT1 mediates the transport of monosaccharides into the vacuole34. In grape, VvTMT1 is significantly expressed in the fruit and is associated with glucose and fructose accumulation during fruit development57. We found that MdTMT1 has the same transmembrane domain structure as its homologs in grape and Arabidopsis (Fig. 5a) and is also localized to the tonoplast (Fig. 5c). Overexpression of MdTMT1 in the tmt1 mutant of Arabidopsis restored the contents of glucose and fructose in leaves (Fig. 6), with similar results observed in apple callus (Fig. 7). Interestingly, silencing of MdTMT1 in the red-fleshed calli reduced the contents of glucose and fructose but not that of sucrose (Fig. 7c).
A comparative proteomic analysis between the calli overexpressing GFP and those overexpressing MdMYB6-GFP revealed a significant increase in the MdTMT1 protein levels in the calli overexpressing MdMYB6-GFP (Fig. 3). GUS reporter analysis, Y1H, ChIP-PCR and EMSA showed that MdMYB6 binds to the MYBCORE element of the MdTMT1 promoter, resulting in increased expression of MdTMT1 (Fig. 8d–g). The transcript level of MdTMT1 and the contents of glucose and fructose were higher in the calli co-overexpressing MdMYB6 and MdTMT1 than in the calli overexpressing MdTMT1 alone (Fig. 7b, c). Together, these results suggest that MdMYB6 promotes the expression of MdTMT1, which transports glucose and fructose into the vacuole, thereby increasing their contents in the callus. However, MdMYB6 was a repressor, and there was no mechanism to show how it could act as an activator to promote the expression of MdTMT1, which might be why MdMYB6 inhibited a repressor of MdTMT1 that would indirectly lead to activation.
MdTMT1 may decrease anthocyanin content by reducing the levels of substrates for anthocyanin biosynthesis
Anthocyanidins are synthesized from leucoanthocyanidin in a reaction catalyzed by anthocyanidin synthase (ANS). Although these intermediate compounds are unstable, UDP-sugar:flavonoid 3-O-glycosyltransferase (UFGT) can rapidly catalyze the conversion of anthocyanidins to anthocyanins by adding a UDPG sugar. In some plants, such as Antirrhinum majus, Gentiana triflora, Perilla frutescens, and Vitis vinifera, the preferred acceptor is anthocyanidin, and the donor is generally UDP-glu58, making UDP-glu content an extremely important factor for anthocyanin biosynthesis. The substrate UDP-gal was found to be positively correlated with anthocyanin content42 and is thought to be synthesized from UDP-glu.
Although many studies have focused on the regulation of structural genes that are required for anthocyanin biosynthesis at the transcriptional and protein modification levels, few studies have focused on substrates for anthocyanin synthesis. In our study, overexpression of MdTMT1 in the red-fleshed callus reduced the contents of UDP-glu and UDP-gal (Fig. 9d), and decreased the expression of UGPase, which encodes the enzyme responsible for converting glucose 1-phosphate to UDP-glu (Fig. 9c). Overexpression of MdMYB6 in the calli overexpressing MdTMT1 increased the expression of MdTMT1, which led to reduced UDP-glu and UDP-gal contents (Fig. 9d). This change resulted in decreased contents of UDP-glu and UDP-gal (Fig. 9d) and consequently reduced anthocyanin content (Fig. 9a, b). This result indicated that MdMYB6 could inhibit anthocyanin biosynthesis by regulating MdTMT1 to decrease the contents of UDP-glu and UDP-gal. Overexpression of MdTMT1 in the calli with silenced MdMYB6 also led to decreased anthocyanin content (Fig. 9). These results indicate that the inhibitory effect of MdTMT1 on anthocyanin content is not affected by the presence of endogenous MdMYB6. The mechanism by which MdTMT1 decreases the contents of UDP-glu and UDP-gal is unknown and should be explored in further research.
On the basis of the above results, we propose a regulatory network of anthocyanin biosynthesis and the transport of monosaccharides across the tonoplast, which are controlled by MdMYB6 and MdTMT1 (Fig. 10). As shown in our analyses, MdMYB6 can directly bind to the promoters of MdANS and MdGSTF12 and reduce their expression, thereby inhibiting the biosynthesis of anthocyanin. Furthermore, MdTMT1 decreases the contents of UDP-glu and UDP-gal, leading to a reduction in anthocyanin content. MdTMT1, which is located at the tonoplast, also increases the contents of glucose and fructose in plant cells, while MdMYB6 can bind to the MdTMT1 promoter and drive increased MdTMT1 expression. These results significantly expand the current understanding of the relationship between sugar levels and anthocyanin contents in apple.
Materials and Methods
Plant materials and processing
‘Purple 3’ and ‘Hongcui 1’ apples were selected from first-generation hybrids created by crossing Malus sieversii f. niedzwetzkyana with Malus × domestica cv. Fuji. The flesh of individual ‘Hongcui 1’ apples was collected five times during fruit development at 60, 80, 100, 115, and 130 days after flowering. The collected tissue was then frozen in liquid nitrogen before storage at −80 °C. Red-fleshed apple callus was created from the young leaves of the ‘Purple 3’ apples, as described by Ji et al59. The red-fleshed callus used for genetic transformation was cultured in Murashige and Skoog medium (MS), which was supplemented with a mixture of 1 mg/L 6-benzylaminopurine and 0.3 mg/L 1-naphthylacetic acid, for 2 weeks. The Arabidopsis tmt1 mutant (stock code N874344) was purchased from http://arabidopsis.info/.
Anthocyanin extraction and quantification
Anthocyanin was extracted as previously described by Jin et al.53, with slight alterations. One gram of plant tissue was first ground to powder in liquid nitrogen, followed by extraction with a solution of 40 mL of 1% (v/v) HCl-methanol at 4 °C in the dark for 24 h. The resulting solution was then centrifuged at 12,000×g for 10 min. An ultraviolet spectrophotometer was then used to determine the supernatant absorbance at 530 nm.
Extraction of total RNA and qRT-PCR analyses
RNA was extracted via a kit (TransGen Biotech, Beijing, China), as described by Xu et al19. The qRT-PCR analyses were carried out on the Bole CFX96 system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s recommended protocols. Each sample was analyzed in triplicate, with MdActin used as the internal reference gene for each quantification. The Ct values were read under default conditions, and the 2−ΔΔCT method was used for data analysis60.
Determination of sucrose, glucose and fructose contents
According to the method of Zhao et al.61, 5 g of tissue was ground into a homogenate and transferred to a 30-mL centrifuge tube. After addition of 25 mL of ultrapure water, the mixture was subjected to ultrasonic extraction in an 80 °C water bath for 1 h, cooled, and then centrifuged for 15 min at 10,000×g. The supernatant was then filtered into a 50-mL volumetric flask, and the residue was added to 15 mL of ultrapure water for extraction. The supernatant volume was adjusted to 50 mL using ultrapure water. The sample was then filtered through a 0.22-μm filter before being assessed via high-performance liquid chromatography (HPLC). The contents of sucrose, glucose, and fructose were calculated according to standard curves using the peak areas of the samples. HPLC was performed using a Shimadzu RID-10A Refractive Index Detector and a YMC Polyamine II chromatographic column (length × inner diameter: 250 × 4.6 mm2, particle diameter: 5 μm); the mobile phase was comprised of acetonitrile:water = 75:25 (v/v); the sensitivity was 4, the injection volume was 10 μL, and the flow rate was 0.8 mL/min at 30 °C.
Determination of UDP-galactose and UDP-glucose contents
The quantities of UDP-gal and UDP-glu were determined as described by Ban et al.42, with slight modifications. Each 5 g sample was ground to powder in liquid nitrogen. One milliliter of 0.5 M perchloric acid was added, and the mixture was kept on ice for 2 min. The samples were then centrifuged at 5000 × g for 10 min at 0 °C. The supernatant was taken to pH 6.5 by adding cooled 2.5 M potassium hydroxide in 1.5 M dipotassium hydrogen phosphate. The resulting solution was then centrifuged at 5000×g for 5 min at 0 °C to remove the potassium perchlorate precipitate. The supernatant was filtered through a 0.45-μm filter before analysis via HPLC. An L-7100 (Hitachi, Tokyo, Japan) was used for the HPLC pump, with an L-7420 (Hitachi) as the UV-vis detector. A D-7500 (Hitachi) was utilized for the integrator, and an L-7300 (Hitachi) was employed for the column oven. The column was a Supelcosil LC-18T with a 4.6 mm inner diameter × 150 mm (Supelco, Bad Homburg, Germany). Solvent A (100 mM dipotassium hydrogen phosphate, 100 mM potassium dihydrogen phosphate and 8 mM tetrabutylammonium hydrogen sulfate) was adjusted to pH 5.3 via the addition of phosphoric acid. Solvent B consisted of a mixture of 70% solvent A and 30% methanol. The gradient system was as follows: 100% solvent A for 2.5 min, 0–40% solvent B for 14 min, 40–100% solvent B for 1 min, 100% solvent B for 6 min, and 100–0% solvent B for 1 min. The flow rate was set to 1.5 mL/min at a temperature of 30 °C. The eluate was measured at an absorbance of 254 nm, and the peaks were identified based on a comparison to standard retention times.
Transformation of the red-fleshed apple callus with MdMYB6 and MdTMT1
The red-fleshed apple callus was transformed as described previously19. The coding sequences (CDSs) of MdMYB6 and MdTMT1 were ligated into the pRI-101 vector containing GFP and the 35S CaMV promoter, respectively, to generate the 35S:MdMYB6-GFP and 35 S:MdTMT1-GFP plasmids (Supplementary Fig. S1A). The CDS of MdMYB6 was ligated into the pCAMBIA1301 vector, which contained a sequence encoding a His tag as well as the 35S CaMV promoter, to construct the 35S:MdMYB6-His recombinant plasmid (Supplementary Fig. S1E). The recombinant plasmids were transformed into Agrobacterium tumefaciens LBA4404. We then infected the red-fleshed calli with the transformed Agrobacterium and cocultured them on MS medium in the dark at 24 °C for 48 h. The resulting calli were then moved onto a screening medium containing 250 mg/L carbenicillin and 50 mg/L kanamycin (Solarbio, Beijing, China) for the MdMYB6- or MdTMT1-overexpressing calli. The screening medium for cultivating the calli cotransfected with MdMYB6 and MdTMT1 contained 250 mg/L carbenicillin, 20 mg/L hygromycin and 50 mg/L kanamycin.
RNA interference to silence MdMYB6 and MdTMT1 in the red-fleshed callus
A 317-bp sequence of MdMYB6 and a 388-bp sequence of MdTMT1 were used to construct the MdMYB6 and MdTMT1 RNAi plasmids. Either MdMYB6ps (MdMYB6 partial sequence) or MdTMT1ps (MdTMT1 partial sequence) was connected to the PFGC-5941 vector containing a chsA intron and the 35 S CaMV promoter in the forward and reverse directions, respectively (see Supplementary Fig. S1F,G). The forward MdMYB6 and MdTMT1 partial sequences were named fMdMYB6ps and fMdTMT1ps, respectively. The reverse MdMYB6 and MdTMT1 partial sequences were named rMdMYB6ps and rMdTMT1ps, respectively. The red-fleshed callus was transformed with these constructs as described above. The calli were screened on medium that contained 20 mg/L hygromycin and 250 mg/L carbenicillin.
Ectopic expression of MdTMT1 in the Arabidopsis tmt1 mutant
Ectopic expression was carried out as described by Wang et al.62, with slight alterations. The recombinant plasmid containing MdTMT1 (Supplementary Fig. S1E) was transformed into A. tumefaciens GV3101, which was utilized to infect the Arabidopsis tmt1 mutant. The T1 transgenic Arabidopsis plants were then selected by plating onto MS medium containing hygromycin. The hygromycin-resistant seedlings were then moved to soil and grown in a growth chamber (Ningbo-Jiangnan, http://www.nbjnyq.com/). The T2 seeds were then collected and grown as described above, and their soluble sugar contents were determined.
Protoplast preparation and analysis of subcellular localization
Protoplasts were prepared as described previously by Wang et al.47, with slight changes. Two grams of callus containing GFP-tagged OE-MdTMT1 was added to 10 mL of a cell wall lysis solution and then placed under a vacuum for 30 min. This process was followed by incubation in the dark at 24 °C for a total of 12 h. Following incubation, the cell suspension was gently mixed to free the protoplasts before 10 mL of W5 solution was added. Nylon fabric (75 μm pore diameter) was then used to filter the solution, followed by centrifugation at 5000 × g. After removal of the supernatant, the protoplasts were suspended in 10 mL of W5 solution and chilled on ice for 30 min. The supernatant was then removed, and the protoplasts were resuspended in 2 mL of MMG solution. An epifluorescence microscope was then used to measure the fluorescence of the protoplasts.
Yeast one-hybrid (Y1H) analysis
The Y1H assays were performed as described by Xu et al.19, with minor alterations. The yeast strain Y187 (Clontech, Palo Alto, CA, USA) was utilized following the manufacturer’s recommended protocols. The CDS of MdMYB6 was ligated into the pGADT7 vector (Supplementary Fig. S1B), while the promoters of MdANS, MdGSTF12 and MdTMT1 were ligated into a pHIS2 vector (Supplementary Fig. S1C,D,I). Different combinations of constructs were then cotransformed into Y187, and the interactions were determined on media with optimal 3-AT concentrations that were deficient in Trp, Leu, and His (SD/-Trp-Leu-His).
GUS staining and activity analyses
The MdTMT1 promoter was ligated to the pCAMBIA1301 vector, which encodes the GUS protein (described in Supplementary Fig. S1H). The constructed MdTMT1 pro:GUS vector was transferred into the WT and OE-MdMYB6 calli. The expression of GUS was detected by histochemical staining as described by Jefferson et al63. Two-week-old calli were submerged in GUS staining buffer, placed under a vacuum for 5 min, and then incubated overnight at 37 °C. The samples were then photographed. For analysis of GUS enzyme activity, 0.5 g of transgenic callus was extracted in 1 mL of GUS extraction buffer. The concentration of the resulting protein sample was then assessed via the Bradford method64. GUS activity was then quantified as described previously22.
Electrophoretic mobility shift assays (EMSAs)
The EMSAs were conducted using an EMSA kit (Pierce, Rockford, IL, USA) following the manufacturer’s recommended protocols. The promoter sequences of MdANS, MdGSTF12, and MdTMT1 were used to create probes labeled with biotin (Sangon Biotech, Shanghai, China). Fifty nanograms of MdMYB6-His DNA was purified for use in binding reactions, which were carried out in 20-mL reaction mixtures containing 17% glycerol, 0.1 mM ethylenediaminetetraacetic acid, 4 mg poly(dI-dC), 100 mM KCl, 1 mM DTT, 25 mM HEPES-KOH (pH 7.5), competitor DNA (25, 50, or 100 pmol), 1 pmol labeled probe, and 50 ng of purified protein. The reactions were then incubated at room temperature for 30 min. The samples were analyzed by electrophoresis on 6% acrylamide gels containing 3.6% glycerol and 0.5% TBE buffer for 2 h at 4 °C after a prerun in 0.5% TBE buffer at 100 V for 1 h. The DNA was then transferred onto nylon membranes, and the signal was detected by chemiluminescent nucleic acid detection.
Chromatin immunoprecipitation-PCR (ChIP-PCR) analyses
The ChIP assays were carried out as described by He et al.65, with slight changes. A ChIP Kit (Upstate, Lake Placid, NY, USA) was used for crosslinking, removal of crosslinking, immunoprecipitation with a GFP antibody (Abmart, Shanghai, China) and elution. The quantity of the immunoprecipitated chromatin was then assessed via PCR. Each analysis was performed in triplicate.
Data analysis, protein sequence alignment and phylogenetic tree construction
All results shown represent an average of three replicates. Significant differences between groups were determined by Duncan’s new multiple range test, with significance tests denoted as i, ii, iii, and iv. Different lowercase letters above columns indicate significant differences (P < 0.05). The phylogenetic tree was constructed with MEGA 5.0, and related protein sequences were analyzed with Clustal X.
References
Zhang, X. Y., Zhang, S. L., Luo, J., Ye, Z. W. & Li, S. C. Advances in research on fruit anthocyanin synthesis. J. Fruit Sci. 21, 456–460 (2004).
Zhang, W. S. et al. Bioactive components and antioxidant capacity of Chinese bayberry (Myrica rubra Sieb. and Zucc.) fruit in relation to fruit maturity and postharvest storage. Eur. Food Res. Technol. 227, 1091–1097 (2008).
Sun, C. D., Huang, H. Z., Xu, C. J., Li, X. & Chen, K. S. Biological activities of extracts from Chinese bayberry (Myrica rubra Sieb. et Zucc.): A review. Plant Foods Hum. Nutr. 68, 97–106 (2013).
Kondo, S., Hiraoka, K., Kobayashi, S., Honda, C. & Terahara, N. Changes in the expression of anthocyanin biosynthetic genes during apple development. J. Am. Soc. Hortic. Sci. 127, 971–976 (2002).
Kim, S. Y., Lee, J. R. & Kim, S. R. Characterization of an Apple anthocyanidin synthase gene in transgenic Tobacco. J. Plant Biol. 49, 326–330 (2006).
Baudry, A. et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, 366–380 (2004).
Ballester, A. R. et al. Biochemical molecular analysis of pink tomatoes, deregulated expression of the gene encoding transcription factor SlMYB12 leads to pink tomato fruit color. Plant Physiol. 152, 71–84 (2010).
Albert, N. W. et al. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784 (2011).
An, X. H., Tian, Y., Chen, K. Q., Wang, X. F. & Hao, Y. J. The apple WD40 protein MdTTG1 interacts with bHLH but not MYB protein to regulate anthocyanin accumulation. Plant Physiol. 169, 710–717 (2012).
Azuma, A. et al. Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor. Appl. Genet 117, 1009–1019 (2008).
Wang, L. K. et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10, 50 (2010).
Feng, S. Q., Wang, Y. L., Song, Y., Xu, Y. T. & Chen, X. S. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta 232, 245–255 (2010).
Espley, R. V. et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor MdMYB10. Plant J. 49, 414–427 (2007).
Espley, R. V. et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 21, 168–183 (2009).
Chagné, D., Lin-Wang, K. & Espley, R. V. et al. An Ancient Duplication of Apple MYB Transcription Factors Is Responsible for Novel RedFruit-Flesh Phenotypes. Plant Physiol. 161, 225–239 (2013).
Aharoni, A. et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332 (2001).
Albert, N. W. et al. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell 26, 962–980 (2014).
Gao, J. J. et al. The myb transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis. Plant Cell Tiss. Organ Cult. 106, 235–242 (2011).
Xu, H. F. et al. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 94, 149–165 (2017).
Koes, R., Verweij, W. & Quattrocchio, F. Flavonoids, a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 10, 236–242 (2005).
Solfanelli, C., Bartolini, S., Vitagliano, C. & Lorenzi, R. Immunolocalization and quantification of IAA after self- and freepollination in Olea europaea L. Sci. Hortic. 110, 345–351 (2006).
Xie, X. B. et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apple. Plant Cell Environ. 35, 1884–1897 (2012).
Bai, S. L. et al. An apple B-box protein, MdCOL11, is involved in UV-Band temperature-induced anthocyanin biosynthesis. Planta 240, 1051–1062 (2014).
Fang H. C., et al.The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 42, 2090–2092 (2019).
An, X. H. et al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 56, 650–662 (2014).
Sun, Y., Li, H. & Huang, J. R. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 5, 387–400 (2012).
Jiang, S. H. et al. MdGSTF6, activated by MdMYB1, plays an essential role in anthocyanin accumulation in apple. Horticulture Res. 6, 40 (2019).
Ruan, Y. L. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67 (2014).
Rae, A., Perroux, J. M. & Grof, C. P. L. Sucrose partitioning between vascular bundles and storage parenchyma in the sugarcane stem: apotential role for the ShSUT1 sucrose transporter. Planta 220, 817–825 (2005).
Sauer, N. Molecular physiology of higher plant sucrose transporters. FEBS Lett. 581, 2309–2317 (2007).
Kühn, C. in Transporters and Pumps in Plant Signaling (eds Geisler, M. & Venema, K.) (Springer-Verlag, Berlin, Heidelberg, 2011) 225–251.
Geiger, D. Plant sucrose transporters from a biophysical point of view. Mol. Plant 4, 395–40 (2011).
Enlder, A. et al. Identification of a Vacuolar Sucrose Transporter in Barley and Arabidopsis Mesophyll Cells by a Tonoplast Proteomic Approach. Plant Physiol. 141, 196–207 (2006).
Wormit, A. et al. Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18, 3476–3490 (2006).
Yamada, K. et al. Monosaccharide absorption activity of Arabidopsis roots depends on expression profiles of transporter genes under high salinity conditions. J. Biol. Chem. 286, 43577–43586 (2011).
Poschet, G. et al. A novel Arabidopsis vacuolar glucose exporter is involved in cellular sugar homeostasis and affects the composition of seed storage compounds. Plant Physiol. 157, 1664–1676 (2011).
Wingenter, K. et al. Increased activity of the vacuolar monosaccharide transporter TMT1 alters cellular sugar partitioning, sugar signaling, and seed yield in Arabidopsis. Plant Physiol. 154, 665–677 (2010).
Schulze, W. X., Schneider, T., Starck, S., Martinoia, E. & Trentmann, O. Cold acclimation induces changes in Arabidopsis tonoplast protein abundance and activity and alters phosphorylation of tonoplast monosaccharide transporters. Plant J. 69, 529–541 (2012).
Weiss, D. Regulation of flower pigmentation and growth: multiple signaling pathways control anthocyanin synthesis in expanding petals. Physiol. Plant 110, 152–157 (2000).
Zheng, Y. J. et al. Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regul. 58, 251–260 (2009).
Sivitz, A. B., Reinders, A. & Ward, J. M. Arabidopsis Sucrose Transporter AtSUC1 Is Important for Pollen Germination and Sucrose-Induced Anthocyanin Accumulation. Plant Physiol. 147, 92–100 (2008).
Ban, Y. S. et al. UDP-sugar biosynthetic pathway: contribution to cyanidin 3-galactoside biosynthesis in apple skin. Planta 230, 871–881 (2009).
Song, Z., Li, T. Z. & Xu, G. X. Studies on the relationship among the anthocyanin sugar and related enzymes activity during the coloring stage of’Fuji’apple. Chin. Agric. Sci. Bull. 24, 255–260 (2008). (in Chinese).
Sainz, M. B., Grotewold, E. & Chandler, V. L. Evidence for direct activation of an anthocyanin promoter by the maize C1 protein and comparison of DNA binding by related Myb domain proteins. Plant Cell 9, 611–625 (1997).
Solano, R. N. C., Avila, J., Canas, L., Diaz, I. & Paz-Ares, J. Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14, 1773–1784 (1995).
Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R. & Weisshaar, B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171 (2005).
Wang, N. et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 96, 39–55 (2018).
Wang, K. L. et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell &. Environment 34, 1176–1190 (2011).
Colquhoun, T. A. et al. PhMYB4 fine-tunes the floral volatile signature of Petunia x hybrida through PhC4H. J. Exp. Bot. 62, 1133–1143 (2011).
Cavallini, E. et al. The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470 (2015).
Paolocci, F. et al. The strawberry transcription factor FaMYB1 inhibits the biosynthesis of proanthocyanidins in Lotus corniculatus leaves. J. Exp. Bot. 62, 1189–1200 (2011).
Xu, H. F. et al. Overexpression of a repressor MdMYB15L negatively regulates anthocyanin and cold tolerance in red-fleshed callus. Biochem. Biophys. Res. Commun. 500, 405–410 (2018).
Jin, H. L. et al. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161 (2000).
Hu, B. et al. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 35, 831–843 (2016).
Martinoia, E., Meyer, S., De Angeli, A. & Nagy, R. Vacuolar transporters in their physiological context. Annu. Rev. Plant Biol. 63, 183–213 (2012).
Schulz, A. et al. Proton-driven sucrose symport and antiport are provided by the vacuolar transporters SUC4 and TMT1/2. Plant J. 68, 129–136 (2011).
Afoufa-Bastien, D. et al. The Vitis vinifera sugar transporter gene family: Phylogenetic overview and macroarray expression profiling. BMC Plant Biol. 10, 22 (2010).
Vogt, T. & Jones, P. Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 1360–1385 (2000).
Ji, X. H. et al. Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f.niedzwetzkyana). Plant Cell Tiss. Organ Cult. 120, 325–337 (2015).
Kenneth, J. L. & Thomas, D. S. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Zhao, Z. Z., Zhang, S. L., Xu, C. J., Chen, K. S. & Liu, S. T. Roles of activities of sucrose-metabolizing enzymes in accumulation of sugars in satsuma mandarin fruit. Acta Hortic. Sin. 28, 112–118 (2001). (in Chinese).
Wang, N. et al. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana). Plant J. 90, 276–292 (2017).
Jefferson, R. A., Kavanagh, T. A. & Bevan, M. W. GUS fusions, beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
He, J. X. et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638 (2005).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31730080; No. 31572091), the Ministry of Science and Technology Key Research and Development Plan (No. SQ2016YFSF030011), and the Shandong Provincial Agricultural Variety Project (2019LZGC007). We thank Dr. Jennifer Smith, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
X.C., N.W. and H.X. conceived and designed the experimental work. H.X. and Q.Z. carried out the experiments. H.X. performed the data analysis. Y.W., S.J., H.F., J.Z., G.Y., and Z.Z. contributed reagents/materials/analysis tools. H.X. and X.C. wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, H., Zou, Q., Yang, G. et al. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic Res 7, 72 (2020). https://doi.org/10.1038/s41438-020-0294-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41438-020-0294-4
This article is cited by
-
The R2R3-MYB transcription factor ZeMYB32 negatively regulates anthocyanin biosynthesis in Zinnia elegans
Plant Molecular Biology (2024)
-
StMYB113 Promotes Anthocyanin Biosynthesis in Potato (Solanum tuberosum L.) Désirée Tubers
Potato Research (2024)
-
Integrated metabolome and transcriptome analysis unveils novel pathway involved in the fruit coloration of Nitraria tangutorum Bobr.
BMC Plant Biology (2023)
-
Identification of R2R3-MYB family in blueberry and its potential involvement of anthocyanin biosynthesis in fruits
BMC Genomics (2023)
-
Overexpression of CaMYB78 transcription factor enhances resistance response in chickpea against Fusarium oxysporum and negatively regulates anthocyanin biosynthetic pathway
Protoplasma (2023)