Abstract
Oculo-auriculo-vertebral syndrome (OAVS) is a clinically heterogeneous disorder, with both genetic and environmental contributors. Multiple genes have been associated with OAVS and common molecular pathways, such as retinoic acid and the PAX-SIX-EYA-DACH (PSED) network, are being implicated in the disease pathophysiology. Biallelic homozygous nonsense or hypomorphic missense mutations in PAX1 cause otofaciocervical syndrome type 2 (OTFCS2), a similar but more severe multi-system disorder that can be accompanied by severe combined immunodeficiency due to thymic aplasia. Here we have identified a multi-generational family with mild features of OAVS segregating a heterozygous frameshift in PAX1. The four base duplication is expected to result in nonsense-mediated decay, and therefore cause a null allele. While there was full penetrance of the variant, expressivity of facial and ear features were variable. Our findings indicate there can be monoallelic and biallelic disorders associated with PAX1, and further implicate the PSED network in OAVS.
Similar content being viewed by others
Introduction
Disorders of the first and second branchial arches include Treacher Collins syndrome (TCS; MIM 154500), auricocondylar syndrome (ACS; MIM 602483), mandibulofacial dysostosis, Guion-Almeida type (MIM 610536), branchio-oto-renal syndrome (BOR; MIM 113650, 610896), Stickler syndrome (MIM 108300) and hemifacial microsomia/oculo-auriculo-vertebral spectrum (HFM/OAVS; MIM 164210). These disorders are thought to result from a combination of inadequate migration and formation of facial mesenchyme during early embryonic life [1]. In TCS, ACS, BOR and Stickler syndrome molecular pathways have been established, however for the spectrum of disease encompassed by OAVS, the full molecular etiology of the condition is the subject of ongoing investigation.
OAVS, also known as Goldenhar syndrome or hemifacial/craniofacial microsomia, is a heterogeneous and complex group of disorders [2]. Craniofacial appearances range from subtle facial asymmetry to HFM or orofacial clefts, preauricular skin tags to microtia and anotia, and epibulbar dermoids to micropthalmia and coloboma [3]. Extracranial defects of the cardiac, renal, vertebral and central nervous systems are reported [3,4,5]. This marked phenotypic variability and a lack of consensus on minimum diagnostic criteria has resulted in considerable discrepancy in reported prevalence, but it is generally now accepted at 1 in 5600 [4].
The etiology of OAVS is still largely unknown and felt to be multifactorial due to both environmental (maternal diabetes, antenatal exposure to vasoactive medications, smoking and twinning) and genetic factors during early embryogenesis [4]. With the widespread adoption of exome or genome sequencing, several genes have now been associated with OAVS [6,7,8,9,10,11], although many have only been identified through segregation in multi-generational single families. Most recently, a large exome/genome sequencing effort of trios identified haploinsufficient variants in SF3B2 in multiple families segregating OAVS [12]. Variants were identified in 3% of sporadic and 25% of familial cases, indicating that while SF3B2 genetic alterations are the most common cause of OAVS identified to date, there is wide genetic heterogeneity for this condition.
The PAX-SIX-EYA-DACH (PSED) network is involved in a variety of developmental processes including roles in morphogenesis of the branchial arches, which acts as the developmental basis for many of the clinical features of OAVS. A proposed mechanism for OAVS is that altered signaling networks cause disrupted migration of cranial neural crest cells, which are essential for normal facial mesenchymal tissue development [4].
The PAX family encode nuclear transcription factors involved in embryogenesis in vertebrates, with five out of the nine PAX genes associated with congenital disorders in humans to date. Biallelic hypomorphic or loss of function variants in PAX1 have been described in several families with otofaciocervical syndrome type 2 (OTFCS2; MIM 615560) [13,14,15,16]. In severe cases, patients can also have severe combined immune deficiency, caused by underdevelopment or absence of the thymus [13].
Here we describe a novel dominant PAX1 variant segregating with full penetrance but variable expressivity in a family with OAVS. All family members are affected with mild but definitive features of OAVS including HFM, misshapen ears and preauricular pits. Imaging on the three available family members did not detect any vertebral anomalies. Our findings suggest a novel cause of OAVS and confirm that heterozygous variants in PAX1 can cause a clinically overt genetic disorder.
Subjects and methods
Clinical presentation of family
We present a three-generation pedigree of five individuals with HFM and microtia in keeping with the OAVS spectrum (Table 1). The proband, III.1, presented at age 5 months with right sided HFM, microtia, small superior sinus and right sided conductive hearing loss. Subsequently the family pedigree revealed an affected mother (II.2), maternal grandfather (I.1), maternal uncle (II.3) and cousin (III.2) (Fig. 1A).
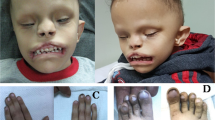
A Pedigree of OAVS features. B Facial features of affected family members with PAX1 variant. III.1 and III.2 display right hemifacial microsomia and microtia, whilst I.1 has subtle facial asymmetry. Misshapen right ears are present in all three individuals ranging from over-folded ear in I.1 to the lobule-type ear in III.2. C Sanger sequencing of PAX1c.1154_1157dup variant. Heterozygous duplication results in overlapping peaks from the frameshift variant site (orange). D PAX1c.1154_1157dup variant located in Exon 4 generates a premature truncation codon in the mRNA transcript (p.Tyr386*) and likely stimulates degradation of the transcript by nonsense-mediated decay (NMD); therefore no protein would be produced from this allele.
Following a normal antenatal course, III.1 was born at term with a birth weight on the 75th centile and was diagnosed with right microtia with a well-formed lobe, residual concha and a small superior sinus at birth (Fig. 1B). The left ear had a cupped and over-folded appearance. Musculoskeletal, cardiovascular and ophthalmological examinations were unremarkable without epibulbar dermoids or vertebral anomalies.
As III.1 grew, a facial asymmetry was noted in keeping with a diagnosis of right hemifacial microtia. He had a normal appearance of his mandible with class I malocclusion. Imaging by CT confirmed an aberrant facial nerve course along with hypoplastic right external auditory canal and middle ear with a dysplastic appearance of his right malleus and incus and an absent stapes. III.1 has moderately severe conductive hearing loss on the right. III.1 attends mainstream school and has no history of developmental delay or intellectual disability. X-ray imaging of the spine revealed no vertebral anomalies.
III.2 was born to non-consanguineous parents after a healthy pregnancy at term with a birth weight on the third centile. III.2 was diagnosed with grade III microtia, lobule-type ear and noted to have a rightward deviated chin and immediately appreciable facial asymmetry in this instance (Fig. 1B). Again musculoskeletal, cardiovascular and ophthalmological examinations were unremarkable. III.2 also had class I malocclusion. III.2 was troubled by a recurrently discharging postauricular sinus throughout his infancy and early childhood. Imaging confirmed a narrow and duplicated internal ear canal accounting for this discharging sinus. In addition III.2 had an absent stapes, and connection to the inner ear. His mastoid was hypoplastic, however appearances of the cochlear and inner ear structures were normal. III.2 also had moderately severe hearing loss on the right. Although III.2 had mildly delayed expressive language this had normalized on school entry with speech and language support and there is no evidence of developmental delay or intellectual disability. X-ray imaging of the spine revealed no vertebral anomalies.
At the initial consultation with III.1 we met I.1 and II.2 who both had subtle facial asymmetry. I.1 had an over-folded right ear and was otherwise well with no significant musculoskeletal, cardiovascular or ophthalmological history, and normal vertebrae on X-ray imaging. II.1 was noted to have a right preauricular pit only and similarly was otherwise well. II.3 has not been formally assessed by our service but is reported to have “small ears and petite facial features”.
Methods
Five members of this family (I.1, II.2, II.4, III.1, III.2) were subjected to exome sequencing using the Agilent V5 capture kit, followed by sequencing on an Illumina HiSeq 4000. Sequencing data were processed by a standard GATK-based pipeline as previously described [17]. Variants were prioritized based on an autosomal dominant inheritance model, starting with variants absent in gnomAD that were of high functional impact.
Sanger sequencing was undertaken to confirm the presence or absence of the PAX1 variant in all family members. Amplification of DNA and sequencing was undertaken using oligonucleotides PAX1ex4F CGTCTCAGTGATGGCGTG and PAX1ex4R CCCAGTACCCTTCCTAACCC, using methodology previously described [17].
Results
Prioritization of high impact variants heterozygous in all affected individuals highlighted a frameshift variant in PAX1; c.1154_1157dup, p.(Tyr386*) (RefSeq: NM_001257096.2), which was absent from gnomAD. Sanger sequencing confirmed segregation within the family (Fig. 1C). The position of the frameshift variant would predict that transcripts harboring this allele would undergo nonsense-mediated decay, and therefore be a null allele (Fig. 1D). This variant can be classified as pathogenic (PVS1, PM2, PP1) using ACMG/AMP criteria [18], although we note it can be difficult to accurately apply such criteria when both monoallelic and biallelic disorders are associated with the same gene, with variable expressivity and penetrance.
Discussion
Biallelic PAX1 mutations are associated with OTFCS2 [13,14,15,16], whereas this family segregates a heterozygous PAX1 variant, with full penetrance but variable expressivity. In comparing the clinical features of this family with OTFCS2, there is phenotypic overlap with this family, of HFM and microtia. There was no evidence of significant developmental delay in this family, nor any vertebral anomalies in the three individuals available for imaging, both of which are observed in patients with biallelic PAX1 variants [3].
A large consanguineous kindred was recently reported where patients were affected with OTFCS2 as well as severe combined immunodeficiency [13], caused by a homozygous nonsense mutation in PAX1. Interestingly, six family members who were heterozygous for the segregating variant were reported to have preauricular pits, although no other phenotypic information was provided. Combined with our findings, this suggests that heterozygous mutations in PAX1 are capable of causing a clinically overt disorder with potentially variable penetrance and expressivity. It remains striking that we observed full penetrance of the duplication variant in this multi-generational family, even with more subtle features. Systematic clinically assessment of PAX1 variant carrier individuals would be valuable to help clinically define the penetrance and expressivity of PAX1 variants, in light of the family presented here.
There is an increasing number of reported genes associated with OAVS, as well as putative risk alleles from GWA studies [19]. A common theme occurring is the involvement of the PSED network, for example, a recurrent missense variant occurring in EYA3 or intragenic genomic alterations of DACH1 or DACH2 [10, 20]. All of these are predicted to act through a dominant genetic mechanism, as opposed to the biallelic loss of function mechanism for PAX1 in OTFCS2.
Many of the genes identified thus far have utilized the multi-generational segregation of OAVS to prioritize variants. Such familial examples also provide insight into the variable penetrance and expressivity of the OAVS phenotypes. As the genetic basis of familial OAVS expands, it will be interesting to learn whether OAVS subcategories could be classified using the observed spectrum of penetrance and expressivity, or on the basis of associated developmental pathways.
Data availability
Variant details are available on ClinVar, accession: SCV002098398.
References
Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–54.
Johnson JM, Moonis G, Green GE, Carmody R, Burbank HN. Syndromes of the first and second branchial arches, part 2: syndromes. Am J Neuroradiol. 2011;32:230–7.
Beleza-Meireles A, Hart R, Clayton-Smith J, Oliveira R, Reis CF, Venancio M, et al. Oculo-auriculo-vertebral spectrum: clinical and molecular analysis of 51 patients. Eur J Med Genet. 2015;58:455–65.
Beleza-Meireles A, Clayton-Smith J, Saraiva JM, Tassabehji M. Oculo-auriculo-vertebral spectrum: a review of the literature and genetic update. J Med Genet. 2014;51:635–45.
Regenbogen L, Godel V, Goya V, Goodman RM. Further evidence for an autosomal dominant form of oculoauriculovertebral dysplasia. Clin Genet. 1982;21:161–7.
Liu Z, Sun H, Dai J, Xue X, Sun J, Wang X. ITPR1 mutation contributes to hemifacial microsomia spectrum. Front Genet. 2021;12:616329.
Lopez E, Berenguer M, Tingaud-Sequeira A, Marlin S, Toutain A, Denoyelle F, et al. Mutations in MYT1, encoding the myelin transcription factor 1, are a rare cause of OAVS. J Med Genet. 2016;53:752–60.
Su PH, Yu JS, Chen JY, Chen SJ, Li SY, Chen HN. Mutations and new polymorphic changes in the TCOF1 gene of patients with oculo-auriculo-vertebral spectrum and Treacher-Collins syndrome. Clin Dysmorphol. 2007;16:261–7.
Tingaud-Sequeira A, Trimouille A, Marlin S, Lopez E, Berenguer M, Gherbi S, et al. Functional and genetic analyses of ZYG11B provide evidences for its involvement in OAVS. Mol Genet Genom Med. 2020;8:e1375.
Tingaud-Sequeira A, Trimouille A, Salaria M, Stapleton R, Claverol S, Plaisant C, et al. A recurrent missense variant in EYA3 gene is associated with oculo-auriculo-vertebral spectrum. Hum Genet. 2021;140:933–44.
Trimouille A, Tingaud-Sequeira A, Lacombe D, Duelund Hjortshoj T, Kreiborg S, Buciek Hove H, et al. Description of a family with X-linked oculo-auriculo-vertebral spectrum associated with polyalanine tract expansion in ZIC3. Clin Genet. 2020;98:384–9.
Timberlake AT, Griffin C, Heike CL, Hing AV, Cunningham ML, Chitayat D, et al. Haploinsufficiency of SF3B2 causes craniofacial microsomia. Nat Commun. 2021;12:4680.
Paganini I, Sestini R, Capone GL, Putignano AL, Contini E, Giotti I, et al. A novel PAX1 null homozygous mutation in autosomal recessive otofaciocervical syndrome associated with severe combined immunodeficiency. Clin Genet. 2017;92:664–8.
Patil SJ, Das Bhowmik A, Bhat V, Satidevi Vineeth V, Vasudevamurthy R, Dalal A. Autosomal recessive otofaciocervical syndrome type 2 with novel homozygous small insertion in PAX1 gene. Am J Med Genet A. 2018;176:1200–6.
Pohl E, Aykut A, Beleggia F, Karaca E, Durmaz B, Keupp K, et al. A hypofunctional PAX1 mutation causes autosomal recessively inherited otofaciocervical syndrome. Hum Genet. 2013;132:1311–20.
Yamazaki Y, Urrutia R, Franco LM, Giliani S, Zhang K, Alazami AM, et al. PAX1 is essential for development and function of the human thymus. Sci Immunol. 2020;5:eaax1036.
Knapp KM, Luu R, Baerenfaenger M, Zijlstra F, Wessels H, Jenkins D, et al. Biallelic variants in SLC35C1 as a cause of isolated short stature with intellectual disability. J Hum Genet. 2020;65:743–50.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Zhang YB, Hu J, Zhang J, Zhou X, Li X, Gu C, et al. Genome-wide association study identifies multiple susceptibility loci for craniofacial microsomia. Nat Commun. 2016;7:10605.
Guida V, Sparascio FP, Bernardini L, Pancheri F, Melis D, Cocciadiferro D, et al. Copy number variation analysis implicates novel pathways in patients with oculo-auriculo-vertebral-spectrum and congenital heart defects. Clin Genet. 2021;100:268–79.
Acknowledgements
The authors thank the family for their involvement and support in this study.
Funding
BJF, LSB and this project were supported by a New Zealand Health Research Council Consolidator Grant. Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
LSB conceived the study. LSB and BJF undertook the molecular analyses. SC and KG coordinated the clinical information. SC and LSB wrote the manuscript. All authors approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study received ethical approval from the New Zealand Health and Disability Committee (16/STH/3). Informed consent was provided for the study, with separate consent obtained for the use of photographs.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carter, S., Fellows, B.J., Gibson, K. et al. Extending the PAX1 spectrum: a dominantly inherited variant causes oculo-auriculo-vertebral syndrome. Eur J Hum Genet 30, 1178–1181 (2022). https://doi.org/10.1038/s41431-022-01154-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-022-01154-2
This article is cited by
-
A Novel Truncating Mutation in PAX1 Gene Causes Otofaciocervical Syndrome Without Immunodeficiency
Journal of Molecular Neuroscience (2023)
-
Happy 30th birthday to the European Journal of Human Genetics!
European Journal of Human Genetics (2022)