Abstract
Congenital autosomal recessive cataract with unknown genetic etiology is one of the most common Mendelian diseases among the Turkic-speaking Yakut population (Eastern Siberia, Russia). To identify the genetic cause of congenital cataract spread in this population, we performed whole-exome sequencing (Illumina NextSeq 500) in one Yakut family with three affected siblings whose parents had preserved vision. We have revealed the novel homozygous c.1621C>T transition leading to premature stop codon p.(Gln541*) in exon 8 of the FYCO1 gene (NM_024513.4). Subsequent screening of c.1621C>T p.(Gln541*) revealed this variant in a homozygous state in 25 out of 29 Yakut families with congenital cataract (86%). Among 424 healthy individuals from seven populations of Eastern Siberia (Russians, Yakuts, Evenks, Evens, Dolgans, Chukchi, and Yukaghirs), the highest carrier frequency of c.1621C>T p.(Gln541*) was found in the Yakut population (7.9%). DNA samples of 25 homozygous for c.1621C>T p.(Gln541*) patients with congenital cataract and 114 unaffected unrelated individuals without this variant were used for a haplotype analysis based on the genotyping of six STR markers (D3S3512, D3S3685, D3S3582, D3S3561, D3S1289, and D3S3698). The structure of the identified haplotypes indicates a common origin for all of the studied mutant chromosomes bearing c.1621C>T p.(Gln541*). The age of the с.1621C>T p.(Gln541*) founder haplotype was estimated to be approximately 260 ± 65 years (10 generations). These findings characterize Eastern Siberia as the region of the world with the most extensive accumulation of the unique variant c.1621C>T p.(Gln541*) in the FYCO1 gene as a result of the founder effect.
Similar content being viewed by others
Introduction
Congenital cataract is a major cause of vision loss in children worldwide and leads to lens opacity that consequently could cause different vision impairments, including vision loss [1]. Congenital cataract occurs with a frequency of 1–6 cases among 10,000 births [2], and up to 25% of cases are hereditary [2, 3]. Hereditary forms of cataract are heterogeneous, and ~70% of cases are nonsyndromic and only lead to damaged lenses [2, 4]. Most hereditary nonsyndromic types of congenital cataract are autosomal dominant and autosomal recessive, and rare cases are known as X-linked recessive forms [4]. To date, more than 50 different loci for congenital cataract have been identified. Among them, 32 genetic loci are known for autosomal recessive forms of cataract and causative variants have been found in 24 genes: GCNT2 (OMIM 600429), LEMD2 (OMIM 616312), TDRD7 (OMIM 611258), DNMBP (OMIM 611282), GJA8 (OMIM 600897), PITX3 (OMIM 602629), FOXE3 (OMIM 601094), LIM2 (OMIM 154045), BFSP1 (OMIM 603307), CRYAA (OMIM 123580), CRYAB (OMIM 123590), CRYBB1 (OMIM 600929), CRYВВ3 (OMIM 123630), SYPA1L3 (OMIM 616655), EPHA2 (OMIM 176946), LSS (OMIM 600909), RNLS (OMIM 609360), AKR1E2 (OMIM 617451), AGK (OMIM 610345), MIP (OMIM 154050), GJA3 (OMIM 121015), HSF4 (OMIM 602438), LONP1 (OMIM 605490), and FYCO1 (OMIM 610019) [5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28].
In Russia, the first report about genetic cases of congenital cataract was published in 2001, where the heterozygous variant c.741T>C p.(Val247Ile) in the GJA8 gene was found in one Russian family with autosomal dominant zonular pulverulent cataract [29]. Later, a more detailed mutational analysis of congenital cataract was performed in the Bashkortostan Republic (Volga-Ural region of Russia) in 40 patients of different ethnic origins including Russians and indigenous Turkic peoples of Volga-Ural region (Tatars and Bashkirs) [30,31,32]. In this cohort of patients, three different heterozygous variants c.68G>T p.(Arg23Thr), c.179G>A p.(Gly60Asp), and с.133_142delTGGGGGGATG p.(Trp45Serfs*72) in the GJA8 gene were found in three unrelated families [31]; a novel variant с.291С>G p.(His97Gln) in the CRYAA gene was found in two unrelated patients [30]; and a novel variant c.del1126_1139 p.(Asp376Glnfs*69) in the GJA3 gene was detected in one family [32]. All detected variants were found to be cosegregated with cataract in these affected families according to the autosomal dominant type of inheritance. To date, no autosomal recessive forms of congenital cataract in Russia have been described [29,30,31,32]. However, according to epidemiological data, congenital cataract of unknown genetic etiology is one of the most frequent autosomal recessive diseases (1:8257) in the indigenous Turkic-speaking population of Yakuts in the Sakha Republic located in Eastern Siberia [33]. The Yakuts (originally named the Sakha) are the largest indigenous people of Siberia (466492 according to the Russian Census, 2010), and they are characterized by specific anthropological, demographic, linguistic, and historical features indicated by their relationships to nomadic Turkic tribes of South Siberia and Central Asia. The genetic data revealed a relatively small size of the Yakut ancestor population and a strong bottleneck effect in the Yakut paternal lineages (~80% of Y chromosomes of Yakuts belong to one haplogroup—N3) [34]. Marriage traditions and geographical isolation had a significant role in the genetic and demographic history of the Yakut population.
The high frequency of some Mendelian disorders in the Yakut population was found to be a result of the founder effect. Recently, the specific founder variants for some autosomal recessive disorders in the Yakut population were identified: missense variant c.806C>T p.(Pro269Leu) in the CYB5R3 gene for methemoglobinemia type 1 (OMIM 250800) [35], GAA-repeat expansion in intron 1 of the FRDA gene for Friedreich ataxia I (OMIM 229300) [36], missense variant c.5741G>A p.(Arg1914His) in the NBAS (previously NAG) gene for SOPH syndrome (OMIM 614800) [37], insertion c.4582insT p.(Gln1553*) in the CUL7 gene for 3M syndrome (OMIM 273750) [38], and splice site variant c.-23+1G>A in the GJB2 gene for autosomal recessive deafness 1A (OMIM 220290) [39].
The aim of this work was to identify the genetic causes of congenital or early onset cataract found with a high frequency in the Yakut population in Eastern Siberia (Russia).
Materials and methods
Patients
Thirty-two children with congenital cataract from 29 unrelated families (three affected siblings in one family and two affected siblings in another family) were selected from 57 visually impaired and blind students of the special boarding school for blind children in Yakutsk (the Sakha Republic, Russia) (Supplementary Table 1). All affected subjects displayed bilateral symmetrical congenital or early onset nuclear cataract. Data on the onset of the cataract in examined patients are presented in Supplementary Table 2. Some of them were subjected to cataract surgery in their early years of life before this study; thus, pictures of their lenses were not available. Twenty-four patients were Yakuts, one patient was Russian, and four patients had mixed ethnic origins. Genealogical analyses showed that congenital cataract was segregated by autosomal recessive inheritance in at least 7 of 29 examined families, while there were no affected relatives in the other 22 families.
DNA was extracted from the blood leukocyte fraction using the phenol–chloroform method. All examination and testing procedures were conducted after written informed consent obtained from all participants or the legal representatives of minor participants involved in the study. Informed consent was consistent with the Declaration of Helsinki. This study was approved by the local Biomedical Ethics Committee of Federal State Budgetary Scientific Institution “Yakut Science Centre of Complex Medical Problems”, Yakutsk, Russia (Protocol No. 16, April 16, 2015).
Whole-exome sequencing (WES)
We performed WES on an Illumina NextSeq 500 Sequencer (Illumina Inc., USA) in one subject with congenital cataract from the Yakut family with three affected siblings and healthy parents. The analysis was conducted with the paired-ending reading method (2 × 151 bp), and the average cover was not less than 70–100×. For sample preparation, a method of selective capture of DNA regions belonging to the coding regions of human genes was applied. The sequence reads generated from the libraries were filtered for quality, aligned, and mapped to the hg19 human reference genome using the gsNap program. The variant calling process for both indels (insertion/deletions) and single nucleotide variants was performed using the Genome Analysis Tool Kit (GATK, http://www.broadinstitute.org/gatk). The functional relevance of the substitution was predicted by SIFT, PolyPhen2-HDIV, PolyPhen2-HVAR, and LRT. We used the data from the 1000 Genomes, ESP6500 and Exome Aggregation Consortium projects to evaluate the population frequencies of identified variants. To assess the clinical relevance of the identified variants, the OMIM database and specialized databases of particular diseases were used.
Sanger sequencing and PCR-RFLP analysis
The variant c.1621C>T p.(Gln541*) in exon 8 of the FYCO1 gene detected by WES was validated by Sanger sequencing and PCR-RFLP analysis. Amplification of the fragment of the FYCO1 exon 8 (349 bp) was conducted using primers (F) 5′-TTGGCCTGCCGGAGCTCTT-3′ and (R) 5′-CTGCTGGACAGCAGGAGGTC-3′. The PCR products were subjected to sequencing using the same primers on an ABI PRISM 3130XL (Applied Biosystems, USA) in the Genomics Core Facility of Institute of Chemical Biology and Fundamental Medicine, Siberian Branch of the Russian Academy of Sciences, Novosibirsk, Russia. Variations in DNA sequences were identified through comparison with the FYCO1 (FYVE and coiled-coil domain autophagy adapter 1 [Homo sapiens (human)]) gene reference sequences chr3(GRCh37): NG_031955.1, NC_000003.11, NM_024513.4, NP_078789.2, XP_011532413.1 (NCBI, Gene ID: 79443).
The screening of the c.1621C>T p.(Gln541*) variant was performed by PCR-RFLP analysis using mismatch primers (F) 5′-TTGGCCTGCCGGAGCTCTT-3′, (R) 5′-AGTGACCTGGAGGAGCAGAAGAAGCAGCGCATT-3′ (270 bp) and restriction endonuclease PstI (Patent RU#2648464 of 26.03.2018, issued by the Federal Service for Intellectual Property of Russian Federation).
Epidemiological data
The Sakha Republic (Yakutia), which includes 36 administrative districts, is the largest federal subject of the Russian Federation located in Eastern Siberia, with a total area of 3103.2 km2. The data on the population and ethnic composition of each district and city were obtained from the Department of the Federal State Statistics Service in the Sakha Republic (Yakutia). The total population of the Sakha Republic is 958,528 people (64.1% urban population), with a density of 0.31 people per km2. The major ethnic groups are Yakuts (48.6%) and Russians (36.9%). The minor ethnic groups are Ukrainians (2.1%), Evenks (2.1%), and Evens (1.5%). Other ethnic groups account for <1%. The prevalence of congenital cataract caused by the c.1621C>T p.(Gln541*) variant in the FYCO1 gene in the Sakha Republic was counted per 10,000 people.
Carrier frequency of с.1621C>T p.(Gln541*)
Screening of the variant c.1621C>T p.(Gln541*) in the FYCO1 gene was performed using the PCR-RFLP method (PstI) in 424 DNA samples of unrelated adult individuals without visual acuity complaints from seven populations of Eastern Siberia: Russians (n = 101), Yakuts (n = 126), Evenks (n = 58), Evens (n = 50), Dolgans (n = 35), Yukaghirs (n = 36), and Chukchi (n = 18). These DNA samples were obtained from the DNA Bank of the Department of Molecular Genetics of Yakut Science Centre of Complex Medical Problems (Yakutsk, Russian Federation). The results of this testing remained completely anonymous.
Genotyping of STR markers
The DNA samples of 25 unrelated patients homozygous for c.1621C>T p.(Gln541*) and the DNA samples of 114 unrelated individuals without this variant were used for the haplotype analysis. All patients were Yakuts from various regions of the Sakha Republic. Six STR markers, D3S3512, D3S3685, D3S3582, D3S3561, D3S1289, and D3S3698, were used for the linkage disequilibrium analysis. The total physical size of the FYCO1 gene region flanking the examined markers was ~28 Mb. All STR markers and primers were selected using the appropriate databases Ensembl dated June–July 2016 (http://www.ensembl.org/index.html) and NCBI dated June–July 2016 (http://www.ncbi.nlm.nih.gov/unists/). The amplified products were resolved on a 10% polyacrylamide gel with ethidium bromide staining under ultraviolet light to verify the size.
Age of variant c.1621C>T p.(Gln541*)
The age of the c.1621C>T p.(Gln541*) variant was estimated using the following equation [40]: q = log [1 − Q/1 − Pn]/log (1 − θ), where Q is the observed frequency of the mutant chromosomes not carrying the progenitor marker allele; Pn is the observed frequency of the marker allele in normal chromosomes; q is the number of generations; and θ is the recombination rate calculated from physical distance between marker and investigated variant (under the assumption that 1 cM = 1000 kb).
For estimation of the age of variant c.1621C>T p.(Gln541*), the data on the STR marker D3S3561 (~6.3 Mb from c.1621C>T) were used. The generation time was 25 years.
Haplotype and phylogenetic analysis
Linkage disequilibrium between the alleles of the STR markers was calculated using the formula δ = (Pd − Pn)/(1 − Pn), where δ is a measure of linkage disequilibrium, Pd is the frequency of the associated allele among mutant chromosomes with c.1621C>T, and Pn is the frequency of the same allele among normal chromosomes without c.1621C>T [41].
Statistical evaluation of differences in the allele frequency of studied markers on 50 chromosomes containing variant c.1621C>T and 228 chromosomes without c.1621C>T was performed using the standard χ2 test.
The phylogenetic network of the founder haplotype with c.1621C>T p.(Gln541*) within the ethno-territorial Yakut groups (Central, Vilyuy and Northern) was constructed using the data on three STR markers, D3S3685, D3S3582, and D3S3561, by Network 5.0 software.
Submission to databases
The novel variant NM_024513.4 (FYCO1): c.1621C>T p.(Gln541*) was submitted to ClinVar under accession number: VCV000693984.1 and Variation ID 693984 (https://www.ncbi.nlm.nih.gov/clinvar/variation/693984/).
Results
Identification of variant c.1621C>T p.(Gln541*) in FYCO1
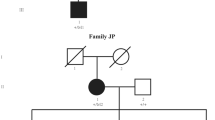
To identify the genetic cause of the congenital cataract cases in the Yakut population, we performed WES (Illumina NextSeq 500) in one affected subject from a Yakut family with three affected siblings and parents with preserved vision. WES revealed a novel homozygous variant c.1621C>T p.(Gln541*) in exon 8 of the FYCO1 gene (FYVE and coiled-coil domain containing 1) (chr3(GRCh37):g.46009205G>A). The FYCO1 gene is located on chromosome 3 (3p21.31) and consists of 18 exons (NM_024513.4). The presence of c.1621C>T p.(Gln541*) was tested by Sanger sequencing and PCR-RFLP analysis in all available members of this Yakut family, and this variant was found in the homozygous state in other affected siblings and in the heterozygous state in their nonaffected parents (Fig. 1).
A Photograph of affected proband P1 (II:3), his nonaffected father F1 (I:1), and his nonaffected mother M (I:2). B Sequencing results: normal subject without с.1621C>T (upper panel), heterozygous for с.1621C>T (middle panel), and homozygous for с.1621C>T (lower panel); C Detection of c.1621C>T in 4% agarose gel by PCR-RFLP analysis (PstI) in examined Yakut family: Mr—marker PUC19/MspI, C1 (control 1)—normal subject (genotype c.[wt];[wt]); F—father of proband I:1 (genotype c.[1621C>T];[wt]); P1, P2 and P3—proband 1 (II:3), proband 2 (II:4) and proband 3 (II:5), respectively (genotype c.[1621C>T];[1621C>T]); M—mother of proband (I:2) (genotype c.[1621C>T];[wt]); C2 (control 2)—normal subject (genotype c.[wt];[wt]); D Pedigree of the Yakut family.
Screening of variant c.1621C>T p.(Gln541*) in FYCO1 in 29 families with congenital cataract
The screening of variant c.1621C>T p.(Gln541*) in all 29 families with congenital cataract was performed by PCR-RFLP analysis and Sanger sequencing. In total, variant с.1621C>T p.(Gln541*) was detected in the homozygous state in 25 affected individuals (25/29, 86%) and in the heterozygous state in one affected individual (1/29, 3%), and it was not found in three families (3/29, 10%) (Fig. 2). Cosegregation of the FYCO1 genotypes with phenotypes of affected and nonaffected relatives was observed in the pedigrees of 10 out of 25 families (Supplementary Fig. 1).
Distribution of congenital cataract caused by homozygous variant c.1621C>T p.(Gln541*) in the FYCO1 gene in the Sakha Republic
The distribution of the homozygous variant c.1621C>T p.(Gln541*) in the FYCO1 gene in the Sakha Republic is shown in Fig. 3. The average rate of congenital cataract caused by the homozygous variant c.1621C>T p.(Gln541*) was 0.29 ± 0.05 per 10,000, with the highest prevalence in the Churapchinsky (1.96 ± 0.98), Ust-Aldansky (1.80 ± 0.90), and Namsky (1.29 ± 0.74) districts of the Sakha Republic (Fig. 3).
The territory of the Sakha Republic is shown in blue (bottom map). The rate of congenital cataract was calculated per 10,000 people, and appropriate data are presented only for the districts and cities of the Sakha Republic with a population of more than 10,000. Complete data are available in Supplementary Table 3.
Carrier frequency of variant c.1621C>T p.(Gln541*) in seven populations of Eastern Siberia
We analyzed the carrier frequency of c.1621C>T p.(Gln541*) in several indigenous populations of the Sakha Republic: Turkic-speaking Yakuts and Dolgans, Tungusic-speaking Evenks, and Evens, Paleo-Asiatic-speaking Chukhi and Uralic-speaking Yukaghirs, as well as Slavic-speaking Russians inhabiting the Sakha Republic (Table 1). Among 424 individuals with normal vision originating from seven studied populations, the c.1621C>T p.(Gln541*) variant was found in a heterozygous state in 12 individuals including Yakuts (10/126, 7.9%), Evenk (1/58, 1.7%) and Even (1/50, 2.0%) and it was absent in Dolgans (0/35), Chukchi (0/18), Yukaghirs (0/36), and Russians (0/101) (Table 1). In addition, the total sample of Yakuts (n = 126) was divided into Central, Vilyuy, and Northern subpopulations according to their ethno-territorial groups. It is noteworthy that the carrier frequency of c.1621C>T p.(Gln541*) obtained for the whole Yakut population (7.9%) varied among the subpopulations of Yakuts, with the highest value (11.1%) found in the central subpopulation of Yakuts (Central Yakuts), a sufficiently lower value (6.4%) found in Vilyuy Yakuts and an undetected value found in Northern Yakuts (Table 1).
Founder haplotype and age of variant с.1621C>T p.(Gln541*)
DNA samples of 25 patients with с.1621C>T p.(Gln541*) in the homozygous state and 114 nonaffected unrelated individuals without this variant were used for the haplotype analysis based on the genotyping of six STR markers (D3S3512, D3S3685, D3S3582, D3S3561, D3S1289, and D3S3698). Linkage disequilibrium was found between the specific alleles of five STR markers (D3S3512, D3S3685, D3S3582, D3S3561, and D3S1289) and variant с.1621C>T p.(Gln541*), while the most distant marker D3S3698 was not in linkage disequilibrium with с.1621C>T p.(Gln541*) (Supplementary Table 4). Based on the χ2 parameter and linkage disequilibrium values, we proposed that the с.1621C>T p.(Gln541*) founder haplotype included the STR alleles D3S3512(6)-D3S3685(4)-D3S3582(3)-D3S3561(4)-D3S1289(3) (Fig. 4). The structure of the identified haplotypes indicates a common origin of all studied mutant chromosomes. The age of the с.1621C>T p.(Gln541*) founder haplotype was estimated at approximately 260 ± 65 years (10 generations); therefore, the beginning of expansion of variant с.1621C>T p.(Gln541*) in the Sakha Republic has been dated to approximately the XVIII century AD.
A Location and physical distance of the STR markers from variant с.1621C>T p.(Gln541*). B STR genotypes of 25 homozygous patients with с.1621C>T p.(Gln541*). The intended founder haplotype for с.1621C>T p.(Gln541*) is marked by a gray color. Group 1—patients who were homozygous for variant с.1621C>T p.(Gln541*) (n = 50 chromosomes); Group 2—Yakut population sample without с.1621C>T p.(Gln541*) (n = 228 chromosomes). (+) and (−) presence and absence of с.1621C>T, respectively, δ: linkage disequilibrium, p: significant differences, q: number of generations.
Phylogenetic analysis of founder haplotype for с.1621C>T p.(Gln541*)
The phylogenetic network of the founder haplotype for с.1621C>T p.(Gln541*) in the FYCO1 gene, which was designed based on the D3S3685, D3S3582, and D3S3561 data, is presented in Supplementary Fig 2. Phylogenetic analysis of the mutant haplotype lineages confirmed the great variety of haplotypes among Central Yakuts. Mutant haplotypes of Vilyuy and Northern Yakuts are probably derived from haplotypes found in the сentral part of the Sakha Republic.
Discussion
In this study, we present the results of a molecular genetic analysis of congenital autosomal recessive cataract, which was registered with high frequency among the Turkic-speaking Yakut population (Eastern Siberia, Russia) [33]. For this analysis, we selected one Yakut family with congenital cataract out of 57 visually impaired and blind students from the special boarding school for blind children (Yakutsk, Russia). This family consisted of three affected siblings and unaffected parents (Fig. 1). In one of three affected siblings, WES revealed a homozygous transition c.1621C>T in exon 8 of the FYCO1 gene located in the CATC2 locus (СTRCT18, OMIM 610019). Subsequent target Sanger sequencing of the FYCO1 gene fragment (exon 8) and PCR-RFLP analysis confirmed the c.1621C>T transition in the homozygous state in all three affected siblings and in the heterozygous state in their healthy parents (Fig. 1).
The c.1621C>T transition leads to premature stop codon p.(Gln541*) (NM_024513.4:c.1621C>T; NP_078789.2:p.Gln541*). This FYCO1 variant has not been previously reported in the 1000 Genomes, ESP6500, and ExAC databases. The size of the FYCO1 (FYVE and coiled-coil domain-containing protein 1) protein is 167 kDa (XP_011532413.1). This protein is associated with the exterior of autophagosomes and mediates microtubule plus-end-directed vesicle transport. The loss of functional activity of FYCO1 inhibits the transport of autophagosomes from the perinuclear region to the periphery of the cell and leads to the accumulation of numerous vesicles, which leads to a loss of transparency of the lens [16, 17]. In 2001, the CATC2 locus was mapped on chromosome 3 in a study of three inbred Arab families with congenital cataract, although genes associated with this form of cataract were not identified [6]. Later, in 2011, a study of 12 Pakistani and one Arab-Israeli family with an autosomal recessive congenital cataract (most families were inbred) identified different variants in the FYCO1 gene located in the critical homozygous region of the CATC2 locus [17]. Recently, several novel variants of the FYCO1 gene were found in British, Saudi Arabian, Egyptian, Pakistani, and Chinese families with autosomal recessive congenital cataract [42,43,44,45,46]. The c.1621C>T nucleotide substitution identified in this study leads to premature stop codon p.(Gln541*) in the functionally significant coiled-coil domain of the FYCO1 protein (NP_078789.2). The schematic structure of the FYCO1 protein and 38 known variants of the FYCO1 gene associated with congenital cataract are presented in Fig. 5. Our results suggest that c.1621C>T p.(Gln541*) is a previously unknown variant that leads to the premature stop codon in the coiled-coil domain and truncates the polypeptide chain of the FYCO1 protein.
A 3D structure of FYCO1. B Schematic organization of the FYCO1 domains: RUN—RUN domain; Сoiled-coil—Сoiled-coil domain; FYVE—FYVE domain; LIR—LC3-interacting region; GOLD—Golgi dynamics domain. C Known variants of FYCO1 associated with congenital cataract. The figure was adapted from PDB ID: 5CX3 and [17]. The clinical significance and other information for variants in the FYCO1 gene associated with congenital cataracts are available in Supplementary Table 5.
Subsequent Sanger sequencing and PCR-RFLP analysis in 29 unrelated families with congenital cataract identified the homozygous variant c.1621C>T p.(Gln541*) in 86% (25/29) of examined individuals. These results suggest that the novel homozygous c.1621C>T p.(Gln541*) variant in the FYCO1 gene is responsible for autosomal recessive cataract CTRCT18 (OMIM 610019) in most affected individuals (86%) in the Yakut population.
Because of the high impact of the novel variant c.1621C>T p.(Gln541*) on congenital cataract among Yakut patients (86%), data on the territorial distribution of this form of disease are very important for the genetic epidemiology of cases of congenital blindness and for estimating the recurrence risks for affected families. We estimated the distribution of the homozygous variant c.1621C>T p.(Gln541*) in the Sakha Republic (average rate was found to be 0.29 ± 0.05 per 10,000). The highest prevalence of congenital cataract caused by c.1621C>T p.(Gln541*) was registered in the central districts of the Sakha Republic: Churapchinsky (1.96 ± 0.98), Ust-Aldansky (1.80 ± 0.90), and Namsky districts (1.29 ± 0.74) (Fig. 3). These results are supported by obtained data on the c.1621C>T p.(Gln541*) carrier frequency. In other indigenous populations of the Sakha Republic, this FYCO1 variant was found among the Tungusic-speaking ethnic groups with a relatively low carrier frequency (2.0% among Evens and 1.7% among Evenks) and was not found in Russians, Yukaghirs, Dolgans, and Chukchi. These results suggest that the ethnic specificity in the distribution of the c.1621C>T p.(Gln541*) variant among the populations of Eastern Siberia is related to the genetic history of the Yakut population. We proposed that variant c.1621C>T p.(Gln541*) in the FYCO1 gene, which causes CTRCT18, could spread in Eastern Siberia as a result of the founder effect.
To test this hypothesis, we performed a haplotype analysis of six polymorphic STR markers on 50 chromosomes with c.1621C>T p.(Gln541*) and 228 chromosomes without c.1621C>T. Out of six analyzed STR markers, linkage disequilibrium was found for five markers (D3S3512, D3S3685, D3S3582, D3S3561, and D3S1289), but not in the most distant marker D3S3698 (Fig. 4). The structure of the identified haplotypes indicates the common origin of all studied mutant chromosomes with c.1621C>T p.(Gln541*) (Fig. 4). Phylogenetic analysis showed that the highest diversity of haplotypes was found in the central subpopulation of Yakuts (Central Yakuts), indicating that the expansion of mutant chromosomes in the territory of the Sakha Republic could start from the Lena-Amga interfluvial area (central part of the Sakha Republic). Mutant haplotypes of Vilyuy and Northern Yakuts are probably derived from haplotypes found in the central part of the Sakha Republic (Supplementary Fig. 2). These data correspond to known historical facts about the initial settling of Yakuts in the central regions and their later expansion to Vilyuy and the northern part of the Sakha Republic [47].
The age of the founder haplotype with variant с.1621C>T p.(Gln541*) was estimated to be ~260 ± 65 years (10 generations). The beginning of the expansion of chromosomes with с.1621C>T p.(Gln541*) in the Sakha Republic has been dated to approximately the XVIII century AD. We compared the age obtained for the founder haplotype with с.1621C>T p.(Gln541*) with relevant data for other variants causing Mendelian disorders found in the Sakha Republic (Table 2). Among eight known Mendelian disorders widely spread in the Sakha Republic, in addition to autosomal recessive cataract 18 (CTRCT18, OMIM 610019), the age of two other founder variants which cause the 3M syndrome 1 (OMIM 273750) and methemoglobinemia due to deficiency of methemoglobin reductase (OMIM 250800) was consistent with this recent historical period (XVII-XVIII centuries AD), which may indicate that some global demographic events occurred at this time, for example, the beginning of Russian colonization of Eastern Siberia (early XVII century AD) [47].
Conclusions
The results of our study suggest that the novel c.1621C>T p.(Gln541*) variant in the FYCO1 gene is responsible for the majority of congenital autosomal recessive cataract cases (86%) in the Yakut population in the Sakha Republic (Eastern Siberia, Russia). We found a high carrier frequency of this FYCO1 variant among the Yakut population (7.9%). The structure of the identified STR haplotypes indicates the common origin of all studied mutant chromosomes with с.1621C>T p.(Gln541*). The beginning of the expansion of chromosomes with с.1621C>T p.(Gln541*) in the Sakha Republic has been dated to approximately the XVIII century AD. These findings characterize Eastern Siberia as the region with highest frequency of the FYCO1 cataract-associated variant c.1621C>T p.(Gln541*) in the world as a result of the founder effect. All obtained data provide important targeted information for genetic counseling of affected Yakut families with congenital cataract and enrich current information on autosomal recessive cataract caused by the FYCO1 gene variants. Nevertheless, further detailed studies of the clinical features of this form of cataract are required.
References
Robinson G, Jan J, Kinnis C. Congenital ocular blindness in children, 1945 to 1984. Am J Dis Child. 1987;141:1321–4. https://doi.org/10.1001/archpedi.1987.04460120087041.
Lambert S, Drack A. Infantile cataracts. Surv Ophthalmol. 1996;40:427–58. https://doi.org/10.1016/S0039-6257(96)82011-X.
Merin S. Inherited cataracts. In: Merin S, editor. Inherited eye diseases. New York: Marcel Dekker, Inc.; 1991.
Shiels A, Bennett T, Hejtmancik J. Cat-Map: putting cataract on the map. Mol Vis. 2010;16:2007–15.
Pras E, Frydman M, Levy-Nissenbaum E, Bakhan T, Raz J, Assia E, et al. A nonsense mutation (W9X) in CRYAA causes autosomal recessive cataract in an inbred Jewish Persian family. Investig Ophthalmol Vis Sci. 2000;41:3511–5.
Pras E, Bakhan T, Levy-Nissenbaum E, Lahat H, Assia E, Garzozi H, et al. A gene causing autosomal recessive cataract maps to the short arm of chromosome 3. Isr Med Assoc J. 2001;3:559–62.
Pras E, Levy-Nissenbaum E, Bakhan T, Lahat H, Assia E, Geffen-Carmi N, et al. A missense mutation in the LIM2 gene is associated with autosomal recessive presenile cataract in an inbred Iraqi Jewish family. Am J Hum Genet. 2002;70:1363–7. https://doi.org/10.1086/340318.
Pras E, Raz J, Yahalom V, Frydman M, Garzozi H, Hejtmancik J, et al. A nonsense mutation in the glucosaminyl (N-acetyl) transferase 2 gene (GCNT2): association with autosomal recessive congenital cataracts. Investig Ophthalmol Vis Sci. 2004;45:1940–5. https://doi.org/10.1167/iovs.03-1117.
Smaoui N, Beltaief O, BenHamed S, M’Rad R, Maazoul F, Ouertani A, et al. A homozygous splice mutation in the HSF4 gene is associated with an autosomal recessive congenital cataract. Investig Ophthalmol Vis Sci. 2004;45:2716–21. https://doi.org/10.1167/iovs.03-1370.
Riazuddin S, Yasmeen A, Yao W, Sergeev Y, Zhang Q, Zulfiqar F, et al. Mutations in beta B3-crystallin associated with autosomal recessive cataract in two Pakistani families. Investig Ophthalmol Vis Sci. 2005;46:2100–6. https://doi.org/10.1167/iovs.04-1481.
Cohen D, Bar-Yosef U, Levy J, Gradstein L, Belfair N, Ofir R, et al. Homozygous CRYBB1 deletion mutation underlies autosomal recessive congenital cataract. Investig Ophthalmol Vis Sci. 2007;48:2208–13. https://doi.org/10.1167/iovs.06-1019.
Ponnam S, Ramesha K, Tejwani S, Matalia J, Kannabiran C. A missense mutation in LIM2 causes autosomal recessive congenital cataract. Mol Vis. 2008;14:1204–8.
Ponnam S, Ramesha K, Tejwani S, Ramamurthy B, Kannabiran C. Mutation of the gap junction protein alpha 8 (GJA8) gene causes autosomal recessive cataract. J Med Genet. 2007;44:e85. https://doi.org/10.1136/jmg.2007.050138.
Ramachandran R, Perumalsamy V, Hejtmancik J. Autosomal recessive juvenile onset cataract associated with mutation in BFSP1. Hum Genet. 2007;121:475–82. https://doi.org/10.1007/s00439-006-0319-6.
Kaul H, Riazuddin S, Shahid M, Kousar S, Butt N, Zafar A, et al. Autosomal recessive congenital cataract linked to EPHA2 in a consanguineous Pakistani family. Mol Vis. 2010;16:511–7.
Pankiv S, Johansen T. FYCO1 linking autophagosomes to microtubule plus end-directing molecular motors. Autophagy. 2010;6:550–2. https://doi.org/10.4161/auto.6.4.11670.
Chen J, Ma Z, Jiao X, Fariss R, Kantorow W, Kantorow M, et al. Mutations in FYCO1 cause autosomal-recessive congenital cataracts. Am J Hum Genet. 2011;88:827–38. https://doi.org/10.1016/j.ajhg.2011.05.008.
Aldahmesh M, Khan A, Mohamed J, Alghamdi M, Alkuraya F. Identification of a truncation mutation of acylglycerol kinase (AGK) gene in a novel autosomal recessive cataract locus. Hum Mutat. 2012;33:960–2. https://doi.org/10.1002/humu.22071.
Evers C, Paramasivam N, Hinderhofer K, Fischer C, Granzow M, Schmidt-Bacher A, et al. SIPA1L3 identified by linkage analysis and whole-exome sequencing as a novel gene for autosomal recessive congenital cataract. Eur J Hum Genet. 2015;23:1627–33.
Ansar M, Chung H, Taylor RL, Nazir A, Imtiaz S, Sarwar MT, et al. Bi-allelic loss-of-function variants in DNMBP cause infantile cataracts. Am J Hum Genet. 2018;103:568–78. https://doi.org/10.1016/j.ajhg.2018.09.004.
Jiao X, Khan SY, Irum B, Khan AO, Wang Q, Kabir F, et al. Missense mutations in CRYAB are liable for recessive congenital cataracts. PLoS ONE. 2015;10:e0137973. https://doi.org/10.1371/journal.pone.0137973.
Khan SY, Vasanth S, Kabir F, Gottsch JD, Khan AO, Chaerkady R, et al. FOXE3 contributes to Peters anomaly through transcriptional regulation of an autophagy-associated protein termed DNAJB1. Nat Commun. 2016;7:10953. https://doi.org/10.1038/ncomms10953.
Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, et al. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–6. https://doi.org/10.1126/science.1195970.
Yasmeen A, Riazuddin SA, Kaul H, Mohsin S, Khan M, Qazi ZA, et al. Autosomal recessive congenital cataract in consanguineous Pakistani families is associated with mutations in GALK1. Mol Vis. 2010;16:682–8.
Zhao L, Chen XJ, Zhu J, Xi YB, Yang X, Hu LD, et al. Lanosterol reverses protein aggregation in cataracts. Nature. 2015;523:607–11. https://doi.org/10.1038/nature14650.
Micheal S, Niewold ITSGl, Siddiqui SN, Zafar SN, Khan MI, Bergen AA. Delineation of novel autosomal recessive mutation in GJA3 and autosomal dominant mutations in GJA8 in Pakistani congenital cataract families. Genes. 2018;9:112. https://doi.org/10.3390/genes9020112.
Chacon-Camacho O, Buentello-Volante B, Velazquez-Montoya R, Ayala-Ramirez R, Zenteno J. Homozygosity mapping identifies a GALK1 mutation as the cause of autosomal recessive congenital cataracts in 4 adult siblings. Gene. 2014;534:218–21. https://doi.org/10.1016/j.gene.2013.10.057.
Patel N, Anand D, Monies D, Maddirevula S, Khan A, Algoufi T, et al. Novel phenotypes and loci identified through clinical genomics approaches to pediatric cataract. Hum Genet. 2017;136:205–25. https://doi.org/10.1007/s00439-016-1747-6.
Polyakov A, Shagina I, Khlebnikova O, Evgrafov O. Mutation in the connexin 50 gene (GJA8) in a Russian family with zonular pulverulent cataract. Clin Genet. 2001;60:476–8. https://doi.org/10.1034/j.1399-0004.2001.600614.x.
Aznabaev MT, Khidiyatova II, Khidiyatova IM, Avkhadeeva SR, Dzhemileva LU, Khusnutdinova EK. [Mutatsii v gene CRYAA - odna iz prichin razvitiya nasledstvennoy vrozhdennoy katarakty]. Meditsinskiy Vestn Bashkortostana. 2014;9:7–9.
Khidiyatova II, Aznabaev MT, Khidiyatova IM, Avkhadeeva SR, Dzhemileva LU, Zinchenko RA, et al. [Analiz gena Konneksina 50 (GJA8) u bol’nykh s nasledstvennoy vrozhdonnoy kataraktoy iz respubliki Bashkortostan]. Meditsinskaya genetika 2014;13:37–42.
Khidiyatova II, Aznabaev MT, Khidiyatova IM, Avkhadeeva SR, Dzhemileva LU, Khusnutdinova EK. [Analiz gena konneksina 46 (GJA3) u bol’nykh nasledstvennoy vrozhdennoy kataraktoy respubliki Bashkortostan]. Meditsinskiy Vestn Bashkortostana. 2016;11:16–20.
Tarskaia L, Zinchenko R, Elchinova G, Egorova A, Korotov M, Basova E, et al. The structure and diversity of hereditary pathology in Sakha Republic (Yakutia). Russ J Genet. 2004;40:1264–72. https://doi.org/10.1023/B:RUGE.0000048669.22362.1c.
Fedorova S, Reidla M, Metspalu E, Metspalu M, Rootsi S, Tambets K et al. Autosomal and uniparental portraits of the native populations of Sakha (Yakutia): implications for the peopling of Northeast Eurasia. BMC Evol Biol. 2013;13. https://doi.org/10.1186/1471-2148-13-127.
Galeeva N, Voevoda M, Spiridonova M, Stepanov V, Polyakov A. Population frequency and age of c.806C>T mutation in CYB5R3 gene as cause of recessive congenital methemoglobinemia in Yakutia. Russ J Genet. 2013;49:457–63. https://doi.org/10.1134/S102279541303006X.
Puzyrev V, Maximova N. Hereditary diseases among Yakuts. Russ J Genet. 2008;44:1141–7. https://doi.org/10.1134/S1022795408100037.
Maksimova N, Hara K, Nikolaeva I, Chun-Feng T, Usui T, Takagi M, et al. Neuroblastoma amplified sequence gene is associated with a novel short stature syndrome characterised by optic nerve atrophy and Pelger-Huet anomaly. J Med Genet. 2010;47:538–48. https://doi.org/10.1136/jmg.2009.074815.
Maksimova N, Hara K, Miyashia A, Nikolaeva I, Shiga A, Nogovicina A, et al. Clinical, molecular and histopathological features of short stature syndrome with novel CUL7 mutation in Yakuts: new population isolate in Asia. J Med Genet. 2007;44:772–8. https://doi.org/10.1136/jmg.2007.051979.
Barashkov NA, Dzhemileva LU, Fedorova SA, Teryutin FM, Posukh OL, Fedotova EE, et al. Autosomal recessive deafness 1A (DFNB1A) in Yakut population isolate in Eastern Siberia: extensive accumulation of the splice site mutation IVS1+1G>A in GJB2 gene as a result of founder effect. J Hum Genet. 2011;56:631–9. https://doi.org/10.1038/jhg.2011.72.
Risch N, de Leon D, Ozelius L, Kramer P, Almasy L, Singer B, et al. Genetic analysis of idiopathic torsion dystonia in Ashkenazi Jews and their recent descent from a small founder population. Nat Genet. 1995;9:152–9. https://doi.org/10.1038/ng0295-152.
Bengtsson B, Thomson G. Measuring the strength of associations between HLA antigens and diseases. Tissue Antigens. 1981;18:356–63. https://doi.org/10.1111/j.1399-0039.1981.tb01404.x.
Khan AO, Aldahmesh MA, Alkuraya FS. Phenotypes of recessive pediatric cataract in a cohort of children with identified homozygous gene mutations (An American Ophthalmological Society Thesis). Trans Am Ophthalmol Soc. 2015;113:T7.
Chen J, Wang Q, Cabrera P, Zhong Z, Sun W, Jiao X, et al. Molecular genetic analysis of Pakistani families with autosomal recessive congenital cataracts by homozygosity screening. Investig Ophthalmol Vis Sci. 2017;58:2207–17. https://doi.org/10.1167/iovs.17-21469.
Li J, Leng Y, Han S, Yan L, Lu C, Luo Y, et al. Clinical and genetic characteristics of Chinese patients with familial or sporadic pediatric cataract. Orphanet J Rare Dis. 2018;13. https://doi.org/10.1186/s13023-018-0828-0.
Iqbal H, Khan SY, Zhou L, Irum B, Ali M, Ahmed MR, et al. Mutations in FYCO1 identified in families with congenital cataracts. Mol Vis. 2020;26:334–44.
Gillespie RL, O’Sullivan J, Ashworth J, Bhaskar S, Williams S, Biswas S, et al. Personalized diagnosis and management of congenital cataract by next-generation sequencing. Ophthalmology. 2014;121:2124–37. https://doi.org/10.1016/j.ophtha.2014.06.006.
Gogolev AI [Etnicheskaya istoriya narodov Yakutii (do nachala XX veka)]. 103. Yakutsk: Izd-vo YAGU; 2004
Marusin A, Kurtanov H, Maksimova N, Swarovsakaja M, Stepanov V. Haplotype analysis of oculopharyngeal muscular dystrophy (OPMD) locus in Yakutia. Russ J Genet. 2016;52:331–8. https://doi.org/10.1134/S1022795416030091.
Swarovskaya M, Stepanova S, Marussin A, Sukhomyasova A, Maximova N, Stepanov V. Genetic variability and structure of SNP haplotypes in the DMPK gene in Yakuts and other ethnic groups of northern Eurasia in relation to myotonic dystrophy. Russ J Genet. 2015;51:619–26. https://doi.org/10.1134/S1022795415060150.
Osakovskiy VL, Goldfarb LG, Platonov FA. [K voprosu proiskhozhdeniya SCA1 mutatsii v yakutskoy populyatsii]. Bulletin’ SO RAMN. 2004;1:103–4.
Acknowledgements
We thank all the patients and blood sample donors who contributed to this study. Special thanks to the physicians and tutors of the special boarding school for blind children (Yakutsk, Russia).
Funding
The study was supported by the Project of the Ministry of Science and Higher Education of the Russian Federation (basic part of funding to M.K. Ammosov North-Eastern Federal University #FSRG-2020–0016) and by the Russian Foundation for Basic Research (grants #18–05–600035_Arctika, #19–34–60023_Perspektiva).
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments and wrote the paper: NAB, FAK, TVB, and FMT; performed the experiments: AVS, VGP, NVS, and LSV; analyzed the data: GPR, NNG, IVM, and AAB; collected the samples: FMT, VGP, FAP, and TEB; discussed and proofed the paper: EKK, OLP, and SAF.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Barashkov, N.A., Konovalov, F.A., Borisova, T.V. et al. Autosomal recessive cataract (CTRCT18) in the Yakut population isolate of Eastern Siberia: a novel founder variant in the FYCO1 gene. Eur J Hum Genet 29, 965–976 (2021). https://doi.org/10.1038/s41431-021-00833-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-021-00833-w
This article is cited by
-
Cataracts in Havanese: genome wide association study reveals two loci associated with posterior polar cataract
Canine Medicine and Genetics (2023)
-
Detection of an FYCO1 nonsense mutation in an affected patient with autosomal recessive cataract (CTRCT18): a case report
Egyptian Journal of Medical Human Genetics (2022)