Abstract
Copy neutral loss of heterozygosity (cnLOH) is a common event in several human malignancies—positing this as a mechanism of carcinogenesis. However, the role of cnLOH in synchronous colorectal cancer (SCRC), a unique CRC subtype, is not well understood. The aim of this study was to establish a cnLOH profile of SCRC using a single-nucleotide polymorphism array (SNP-A), and to explore associations between cnLOH and the genomic landscape of frequently mutated genes in SCRC. Among 74 paired SCRC cases, the most frequently altered regions were 16p11.2–p11.1 (59.5%) and 11p11.2–p11.12 (28.4%). Notably, the 6q11.21–q11.22 region altered by cnLOH was uniquely associated with polyclonal SCRCs (p = 0.038). Together, our analysis suggests that inactivation of tumor suppressor genes and cnLOH are rare events among SCRC cases. This study defines distinct patterns of cnLOH in SCRC, and provides initial evidence of a role for cnLOH in SCRC etiology.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide [1]. Patients with CRC have an increased risk for developing multiple primary CRCs, which encompasses synchronous CRC (SCRC) and metachronous CRC [2]. SCRC cases provide a good model to evaluate a possible field effect—neoplasms that develop in a background of common etiologic factors within an individual, and which may also share molecular features indicating a likely clonal origin [3].
Copy-number alterations (CNAs) are recurrent molecular changes in human cancers and show a tumor-specific landscape of DNA gains and losses [4]. Several studies have posited a possible clonal origin in tumor subsets, including bilateral breast cancer or SCRC cases, based on the concordance of CNA profiles or mutational status [5, 6]. As such, we recently defined a SCRC classification based upon paired-tumor clonality via single-nucleotide polymorphism arrays (SNP-As), which allowed us to characterize SCRC genomic profiles by identifying DNA gains and losses [6]. SCRC cases were classified as monoclonal when paired tumors shared similar patterns of somatic changes indicative that could be derived from a single cell or polyclonal if paired tumors had independent molecular profiles. Combination of clonality analysis and tumor location also led to SCRC categorization as monoclonal monosegmental (MM), monoclonal pancolonic (MP), polyclonal monosegmental (PM), and polyclonal pancolonic (PP) [6].
The use of SNP-A allows for identification of loss of heterozygosity (LOH) and copy neutral LOH (cnLOH) [7]. cnLOH leads to LOH by duplication of one chromosome (or chromosomal region) and concurrent loss of the other allele [8]. According to the two-hit-hypothesis by Knudson, regions with LOH may contain tumor suppressor genes (TSGs), which could be inactivated by either a non-synonymous variant or a genomic loss. In this context, cnLOH could arise as an alternative mechanism to inactivate TSGs as well as activate oncogenes in CRC. Indeed, several studies have described gains of function of homozygous variants in oncogenes and have found that homozygously mutated genes also are localized in cnLOH regions [9].
In this study, we examined regions with cnLOH using SNP-A in a cohort of SCRC patients, and association of cnLOH by clonality and tumor location [6]. We also integrated the mutational profile of frequently mutated genes in SCRCs with the presence of cnLOH in the regions where these genes are encoded in order to explore underlying SCRC etiologies.
Materials and methods
See Supplementary Material Boxes 1 and 2.
Results
cnLOH profiling in SCRC
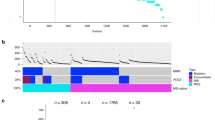
Overall, the majority of SCRC cases showed important proportions of regions affected by cnLOH—with a median of 7.5 events per sample. By categorization, we observed a median of 7, 4.5, 8, and 8 cnLOH events for MM, MP, PM, and PP, respectively (Fig. 1). Overlap between CNAs and regions with LOH were illustrated for each SCRC subtype (Supplementary Material Box 3). Recurrent chromosome regions in our SCRC samples affected by cnLOH are listed in Table 1. Notably, the most commonly altered region across all SCRC subtypes was 16p11.2–p11.1 (59.5%), followed by 11p11.2–p11.12 (28.4%) (Supplementary Material Box 4). Across monoclonal groups, a total of 20 tumors showed cnLOH in 16p11.2–p11.1, including 14 paired tumors from the same patients (70.0%). Twenty tumors also showed the alteration within polyclonal groups, of which only 10 were paired tumors (50.0%). While SCRC groups presented with similar alteration patterns, region 6p22.1–p21.32 affected by cnLOH was uniquely linked to polyclonal groups (p = 0.038) (Table 1B).
cnLOH in CRC-related genes
Recent studies have proposed cnLOH as a “second hit” to inactivate TSGs as well as to activate oncogenes [7, 10]. In order to study cnLOH in SCRC, we explored associations between the presence of cnLOH and genomic landscape of commonly altered SCRC genes. We evaluated DNA sequencing data (where only protein truncating and damaging missense variants were considered) and examined regions where these genes were located to integrate mutation status with cnLOH events in SCRC. In our cohort, 33 (44.6%) tumors presented with APC variants—of which 9 cases (27.3%) also had cnLOH detected. Approximately 37.5% of monoclonal groups presented with both APC variants and cnLOH events in this region, compared with only 18.7% of polyclonal cases. KRAS variants were also observed in 32 (43.2%) neoplasms, irrespective of cnLOH events. Thirteen (17.6%) tumors had detected variants in TP53, with corresponding cnLOH observed in only one (7.7%) of these tumors. Seven (9.5%) tumors presented with FBXW7 variants, although only one (14.3%) variant was associated with cnLOH. Although SMAD4 variants were associated with the MM subgroup (p = 0.049), no tumors presented with concurrent cnLOH (Table 2).
Discussion
SCRC is a heterogeneous disease that remains poorly understood. Consequently, defining genetic patterns specific to SCRC cases may provide critical insight into possible clonal origins of these tumors. Previous work by our group explored CNAs and tumor location for paired SCRCs, and defined a unique classification system based on clonality and tumor site [6]. In this study, we employed SNP-A to assess cnLOH profiles in SCRC among all cases and by subgroup classification in order to better understand etiologies underlying sporadic colorectal carcinogenesis.
Previous reports have analyzed cnLOH profiles across diverse gastrointestinal tumors and revealed the lowest level of regions with cnLOH (3 per sample) for colon cancers by tumor type [10]. Nevertheless, compared with previous results, our specific interrogation of SCRCs revealed a higher frequency of regions altered by cnLOH (median of 7.5 events per sample). We also observed that SCRCs showed a distinct pattern of cnLOH events compared with cases with one primary invasive CRC [10]. It is important to note that while previous studies have leveraged similar techniques and cnLOH criteria, the biospecimen type (e.g. fresh tissue vs FFPE) across studies may also lend to some of these noted differences. Our results also pointed out that the generation of regions with cnLOH in SCRC occurred mainly in the genomic region 16p11.2–p11.1 (59.5%). In contrast, several studies have reported high frequencies of 3p, 8q, 13q, or 20q [11], as well as 5q12.1–q35.3 [10] regions in CRC. Consequently, our findings emphasize the importance of considering CRC heterogeneity in clinical settings and support additional large-scale studies to validate these results. In addition, comparison of cnLOH profiles by clinicopathological features and between SCRC and MCRC cases would provide additional insight into distinct etiologies of colorectal carcinogenesis.
Our discovery of LOH in region 16p11.2–11.1 was largely attributable to cnLOH as it is not a region affected by genomic alterations. Previous studies have reported homologous segmental duplications in this region [12] and, importantly, this region has been associated with early-onset obesity [13] (a well-known risk factor for CRC). Additional studies have revealed that SN2B1 is encoded within this region, and is a gene associated with cancer progression [14]. MAZ—a transcription factor upregulated in several human cancers—is also located in this region [15, 16]. Indeed, several studies have suggested an important role for MAZ in CRC progression by inducing inflammation through the regulation of STAT3 signaling [17]. Additionally, our analysis revealed differential regions altered by cnLOH by SCRC classification, as the 6p22.1–p21.32 genomic region was uniquely associated with the polyclonal tumor subgroup. This finding, together with the more common cnLOH in the 16p11.2–p11.1 region, or the connection of APC variants and cnLOH events in the corresponding region, support our idea that altered chromosomal segments linked with the SCRC carcinogenesis may provide more detailed insight into SCRC clonality [6, 18, 19].
cnLOH events could also act as a “second hit” to achieve biallelic inactivation of TSGs—a potential mechanism for oncogene activation [20]. In this study, we examined the mutational status of APC recurrently inactivated in CRC and its association with cnLOH. Our results showed the possibility that, among SCRC cases, cnLOH could contribute to the functional impairment of this gene, more frequently in monoclonal tumors. However, as no previous associations have been observed between TP53, FBXW7, and SMAD4, with cnLOH, these results suggest that cnLOH events may not play a main role in SCRC. Moreover, cnLOH may not be involved in oncogenic activation of KRAS among these cases—however, prospective studies are warranted to confirm these findings.
In summary, SCRC cases harbor distinct molecular patterns, including the number of cnLOH per tumor as well as regions frequently altered by cnLOH, when compared with single primary invasive CRC profiles. Furthermore, inactivation of some TSGs via damaging variants and cnLOH were rare events among SCRC cases. Additional studies are needed to further explore the role of cnLOH in SCRC etiology.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;1–31.
Lam AKY, Chan SSY, Leung M, Lam AKY, Chan SSY, Leung M. Synchronous colorectal cancer: clinical, pathological and molecular implications. World J Gastroenterol. 2014;20:6815–20.
Lochhead P, Chan AT, Nishihara R, Fuchs CS, Beck AH, Giovannucci E, et al. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol. 2015;28:14–29.
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J. et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905.
Song F, Li X, Song F, Zhao Y, Li H, Zheng H. et al. Comparative genomic analysis reveals bilateral breast cancers are genetically independent. Oncotarget. 2015;6:31820–9.
Perea J, García JL, Corchete L, Lumbreras E, Arriba M, Rueda D, et al. Redefining synchronous colorectal cancers based on tumor clonality. Int J Cancer. 2018;144:1596–608.
Torabi K, Miró R, Fernández-Jiménez N, Quintanilla I, Ramos L, Prat E. et al. Patterns of somatic uniparental disomy identify novel tumor suppressor genes in colorectal cancer. Carcinogenesis. 2015;36:1103–10.
Makishima H, Maciejewski JP. Pathogenesis and consequences of uniparental disomy in cancer. Clin Cancer Res. 2011;17:3913–23.
Tuna M, Smid M, Zhu D, Martens JWM, Amos CI. Association between acquired uniparental disomy and homozygous mutations and HER2/ER/PR status in breast cancer. PLoS ONE. 2010;5:e15094.
Torabi K, Erola P, Alvarez-Mora MI, Díaz-Gay M, Ferrer Q, Castells A, et al. Quantitative analysis of somatically acquired and constitutive uniparental disomy in gastrointestinal cancers. Int J Cancer. 2019;144:513–24.
Andersen CL, Wiuf C, Kruhøffer M, Korsgaard M, Laurberg S, Ørntoft TF. Frequent occurrence of uniparental disomy in colorectal cancer. Carcinogenesis. 2007;28:38–48.
Redaelli S, Maitz S, Crosti F, Sala E, Villa N, Spaccini L, et al. Refining the phenotype of recurrent rearrangements of chromosome 16. Int J Mol Sci. 2019;20:1–17.
Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K. et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70.
Cheng Y, Duan C, Zhang C. New perspective on SH2B1: an accelerator of cancer progression. Biomed Pharmacother. 2020;121:109651.
Maity G, Haque I, Ghosh A, Dhar G, Gupta V, Sarkar S, et al. The MAZ transcription factor is a downstream target of the oncoprotein Cyr61/CCN1 and promotes pancreatic cancer cell invasion via CRAF–ERK signaling. J Biol Chem. 2018;293:4334–49.
Yang Q, Lang C, Wu Z, Dai Y, He S, Guo W, et al. MAZ promotes prostate cancer bone metastasis through transcriptionally activating the KRas-dependent RalGEFs pathway. J Exp Clin Cancer Res. 2019;38:1–17.
Triner D, Castillo C, Hakim JB, Xue X, Greenson JK, Nuñez G, et al. Myc-associated zinc finger protein regulates the proinflammatory response in colitis and colon cancer via STAT3 signaling. Mol Cell Biol. 2018;38:1–15.
Di J, Yang H, Jiang B, Wang Z, Ji J, Su X. Whole exome sequencing reveals intertumor heterogeneity and distinct genetic origins of sporadic synchronous colorectal cancer. Int J Cancer. 2018;142:927–39.
Hänninen UA, Wirta EV, Katainen R, Tanskanen T, Hamberg J, Taipale M, et al. Exome and immune cell score analyses reveal great variation within synchronous primary colorectal cancers. Br J Cancer. 2019;120:922–30.
Sanada M, Suzuki T, Shih LY, Otsu M, Kato M, Yamazaki S. et al. Gain-of-function of mutated C-CBL tumour suppressor in myeloid neoplasms. Nature. 2009;460:904–8.
Acknowledgements
We thank the Tumor Registry of the Pathology Department of the 12 University Hospital, Madrid.
Funding
This work was funded by Projects PI16/01650 to JP, and PI16/01920 to RG-S, from the Spanish Ministry of Health and Consumer Affairs and FEDER, by Project 2012-0036 from the Mutua Madrileña Foundation. ANH was supported by the National Institutes of Health K12 HD043483 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Tapial, S., García, J.L., Corchete, L. et al. Copy neutral loss of heterozygosity (cnLOH) patterns in synchronous colorectal cancer. Eur J Hum Genet 29, 709–713 (2021). https://doi.org/10.1038/s41431-020-00774-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-020-00774-w
This article is cited by
-
P53 loss of heterozygosity (LOH) in formalin-fixed paraffin-embedded leiomyosarcoma (LMS): a novel report
Irish Journal of Medical Science (1971 -) (2024)
-
Loss of heterozygosity related to TMB and TNB may predict PFS for patients with SCLC received the first line setting
Journal of Translational Medicine (2021)