1. Name of the disease (synonyms)
Primary congenital glaucoma (PCG).
Glaucoma, congenital (GLC).
2. OMIM# of the disease
231300- GLC3A.
600975- GLC3B.
613085- GLC3C.
613086- GLC3D.
617272- GLC3E.
3. Name of the analysed genes or DNA/chromosome segments
CYP1B1.
LTBP2.
MYOC.
FOXC1.
TEK.
4. OMIM# of the gene(s)
CYP1B1 MIM# 601771.
LTBP2 MIM# 602091.
MYOC MIM# 601652.
FOXC1 MIM# 601090.
TEK MIM# 600221.
Review of the analytical and clinical validity, as well as of the clinical utility of DNA-based testing for variants in the CYP1B1, LTBP2 and MYOC gene(s) in
⊠ diagnostic, | |||||
⊠ predictive and | |||||
⊠ prenatal settings and for | |||||
⊠ risk assessment in relatives. |
Similar content being viewed by others
1. Disease characteristics
1.1 Name of the disease (synonyms)
Primary congenital glaucoma (PCG).
Glaucoma, Congenital (GLC).
1.2 OMIM# of the disease
231300- GLC3A.
600975- GLC3B.
613085- GLC3C.
613086- GLC3D.
617272- GLC3E.
1.3 Name of the analysed genes or DNA/chromosome segments
1.3.1 Core genes (irrespective if being tested by Sanger sequencing or next-generation sequencing)
CYP1B1.
LTBP2.
MYOC.
FOXC1.
1.4 OMIM# of the gene(s)
CYP1B1 MIM# 601771.
LTBP2 MIM# 602091.
MYOC MIM# 601652.
FOXC1 MIM# 601090.
TEK MIM# 600221.
1.5 Mutational spectrum
Primary congenital glaucoma (PCG) is frequently inherited as an autosomal recessive severe form of glaucoma resulting from obstruction in the aqueous humour drainage due to congenital developmental anomalies in anterior chamber angle structures [1, 2].
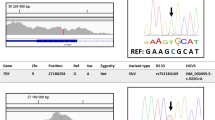
CYP1B1 (NM_000104.3) is located on chromosome 2p21–22 (GLC3A locus, 3 exons) and variants in this gene are the most common cause of PCG. CYP1B1 variants were identified in 41 (75.9%) of 74 Saudi Arabian patients with PCG (54 unrelated families) [3]. A total of 13 variants: 9 missense variants (c.182 G > A [p.(G61E)], c.355 G > T [p.(A119S)], c.1169 G > A [p.(R390H)], c.1310 C > T [p.(P437L)], c.1322 A > G [p.(D441G)], c.1328 C > G [p.(A443G)], c.1396 G > A [p.(G466S)], c.1397 G > A [p.(G466D)], c.1405 C > T [p.(R469W)]), 2 deletions (c.434_443del [p.(R145Pfs*4)] and c.1064_1076del [p.(R355Hfs*69)]), and 2 nonsense variants (c.1063 C > T [p.(R355*)], c.1330 C > T [p.(R444*)]) were identified in these families, and the p.G61E variant was the most common variant detected. PCG cases with a CYP1B1 variant also had higher postoperative visit indices in terms of postoperative haze and the need for anti-glaucoma medications [3].
Variant analysis of the coding exons of CYP1B1 also showed either homozygous or compound heterozygous variants in all 30 affected individuals in 24 of 25 Saudi Arabian families with PCG [4]. Three distinct missense variants were defined in the 24 Saudi Arabian families: c.182 G > A [p.(G61E)], c.1405 C > T [p.(R469W)] and c.1120 G > A [p.(D374N)] in 78%, 10% and 6% of the PCG chromosomes analysed, respectively. The c.−611C > T (rs2567206) promoter variant of CYP1B1 was also strongly associated with PCG in a large cohort of 301 affected individuals, and the intraocular pressure control in patients with CYP1B1 variants was poorer compared with patients who did not harbour any variants [5].
CYP1B1 variants were analysed in a large cohort of European PCG patients composed of 161 unrelated index cases. Approximately 30% of these patients carried recessive disease-causing variants that in most cases led to null CYP1B1 enzymatic activity. Besides, most PCG patients carrying null or putative null genotypes showed severe bilateral phenotypes featured by very early disease onset, quite frequently at birth. The mean number of trabeculectomies per eye was significantly higher in carriers than in non-carriers [6].
Using whole genome exon-focused array comparative genome hybridisation (aCGH), 12 out of 20 Korean PCG patients were identified with at least one rare gene-containing copy number variation (CNV) each, corresponding to 25 CNVs (5 deletions and 20 duplications) at frequencies of 5–30% in PCG patients [7]. CYP1B1 CNV was also analysed by Multiplex Ligation-dependent Probe Amplification (MLPA) in 50 PCG that were recruited through the Australian and New Zealand Registry of Advanced Glaucoma, and no deletions or duplications were found in any of the cases [8].
Variants in LTBP2 (NM_000428.2), which is located on chromosome 14q24.3 (GLC3C chromosomal loci, 36 exons), can cause PCG [9, 10]. The LTBP2 c.895 C > T [p.R299*] variant accounted for 33.8% of disease alleles in 37 PCG patients (34 families) from the Roma/ Gypsy population [11]. Homozygosity for the LTBP2 c.895 C > T [p.R299*] variant was also associated with a more severe clinical phenotype and poorer outcome despite a higher number of surgical interventions [9, 11].
MYOC (NM_000261.1), located on chromosome 1q24 (3 exons), also codes for the myocilin/ trabecular meshwork-induced glucocorticoid response protein and MYOC variants account for 5.5% of PCG cases [12]. A further chromosomal locus has been identified at position 1p36 (GLC3B) but no gene has been identified as yet [13].
1.6 Analytical methods
The oculome exome gene panel contains a sub panel for anterior segment dysgenesis and glaucoma: ADAMTS18, AGBL1, ALDH18A1, ATOH7, B3GLCT, BEST1, BMP7, CHRDL1, CHST6, COL4A1, COL8A2, CRIM1, CRYGC, CYP1B1, DCN, EYA1, FBN1, FOXC1, FOXE3, FOXL2, GJA1, GNPTG, GSN, KERA, KRT12, KRT3, LAMB2, LCAT, LMX1B, LTBP2, MIR184, MYOC, NOTCH2, OPTN, PAX3, PAX6, PEX2, PIKFYVE, PITX2, PITX3, PRDM5, PXDN, RAB18, RAB3GAP1, RAB3GAP2, SEC23A, SH3PXD2B, SIX3, SLC16A12, SLC38A8, SLC4A11, SLC4A4, TACSTD2, TBC1D20, TGFBI, UBIAD1, VSX1, WDR36, ZEB1 genes (http://www.labs.gosh.nhs.uk/media/764794/oculome_v8.pdf). Sanger sequencing is less frequently used to screen genes but is used for validation of identified variants.
There are also human gene variant databases that are available to the public, e.g., LOVD (https://databases.lovd.nl/shared/genes) and Human Genetic Variation Database (http://www.hgvd.genome.med.kyoto-u.ac.jp).
1.7 Analytical validation
Parallel bi-directional fluorescent Sanger sequencing of known controls is required to validate procedures. Diagnostic testing must be carried out within a laboratory environment working to standards compliant with the ISO 15189.
1.8 Estimated frequency of the disease (Incidence at birth (“birth prevalence”) or population prevalence. If known to be variable between ethnic groups, please report)
Prevalence of PCG varies between different ethnic groups and ranges from 1 in 18,500 in the UK [14], 1 in 3300 in Andhra Pradesh (Southern India) [15], 1 in 2500 in the middle-east (Saudi Arabia) [16], and 1 in 1250 in the Rom (Gypsy) population of Slovakia [17]. A higher prevalence is observed in ethnic groups in which parental consanguinity is common [18, 19].
1.9 Diagnostic setting
Yes. | No. | |
A. (Differential) diagnostics | ⊠ | ☐ |
B. Predictive Testing | ⊠ | ☐ |
C. Risk assessment in relatives | ⊠ | ☐ |
D. Prenatal | ⊠ | ☐ |
Comment: Not applicable
2. Test characteristics
Genotype or disease | A: true positives | C: false negative | ||
Present | Absent | B: false positives | D: true negative | |
Test | ||||
Positive | A | B | Sensitivity:specificity: | A/(A+C)D/(D+B) |
Negative | C | D | Pos. predict. value:Neg. predict. value: | A/(A+B)D/(C+D) |
2.1 Analytical sensitivity
(proportion of positive tests if the genotype is present)
2.1.1 If tested by conventional Sanger sequencing
Less than 100% because the primers may be localised on sequences containing SNVs (single nucleotide variants) or rare variants, which results in a preferential amplification of one allele (allele dropout). A supplementary deletion/duplication diagnostic test should be performed for genes with a known proportion of large genomic deletions/duplications.
2.1.2 If tested by next-generation sequencing
Less than 100%. The proportion is likely 100%, because there might be disease-causing variants in regions that could not be enriched and/or sequenced by next-generation sequencing owing to suboptimal coverage of some regions of interest with this technology, but depending on next-generation sequencing strategy. If amplicon-based enrichment strategies are being used, primers may be localised on SNVs or rare variants, which results in preferential amplification of one allele. In patients with a highly suggestive phenotype in whom testing for specific gene alterations proves negative, a supplementary deletion/duplication diagnostic test should be performed for genes with a known proportion of large genomic deletions/duplications.
2.2 Analytical specificity
(proportion of negative tests if the genotype is not present)
2.2.1 If tested by conventional Sanger sequencing
Estimated analytical specificity nearly 100%, based on false positives that may arise due to misinterpretation of rare polymorphic variants that rarely occur in Sanger sequencing.
2.2.2 If tested by next-generation sequencing
Less than 100%. The risk of false positives owing to misinterpretation of rare polymorphic variants may be higher compared with Sanger sequencing because of greater number of analysed genes.
2.3 Clinical sensitivity
(proportion of positive tests if the disease is present)
The clinical sensitivity can be dependent on variable factors like age or family history. In such cases, a general statement should be given, even if a quantification can only be made case by case.
2.3.1 If tested by conventional Sanger sequencing
PCG is a congenital eye defect presenting at less than 6 months of age in most cases and >80% within the first year [20, 21]. PCG occurs more frequently in males than in females [22, 23] and is bilateral in 70 to 80% cases [24, 25]. The manifestations of PCG show a particular concordance in monozygotic twins and discordance in dizygotic twins [18]. The disease classically presents with the symptoms of epiphora (excessive tearing), photophobia (hypersensitivity to light), and blepharospasm (inflammation of the eyelids) [26]. Other clinical features include buphthalmos (enlargement of the eye), corneal oedema, and breaks in Descemet’s membrane (Haab’s striae). The clinical sensitivity is 10–40%.
2.3.2 If tested by next-generation sequencing
See section 2.3.1
2.4 Clinical specificity
(proportion of negative tests if the disease is not present) The clinical specificity can be dependent on variable factors such as age or family history. In such cases, a general statement should be given, even if a quantification can only be made case by case.
2.4.1 If tested by conventional Sanger sequencing
Unknown
2.4.2 If tested by next-generation sequencing
See section 2.4.1.
2.5 Positive clinical predictive value
(life time risk to develop the disease if the test is positive)
The positive clinical predictive value is estimated to be >99% if biallelic variants in CYP1B1 or LTBP2 genes that affect function are present.
2.6 Negative clinical predictive value
(Probability not to develop the disease if the test is negative). Assume an increased risk based on family history for a non-affected person. Allelic and locus heterogeneity may need to be considered.
Index case in that family had been tested:
The mean age of presentation of PCG ranges from 3 to 4 months among Asians, Saudi Arabians and Indians, and up to 11 months in Western countries [27,28,29]. If the index case is asymptomatic by 5 years of age and has a negative test result, it is highly predictive of unaffected status and the negative clinical predictive value will be close to 100%.
Index case in that family had not been tested:
If the index case is asymptomatic by 5 years of age and has a negative test result, it is highly predictive of unaffected status and the negative clinical predictive value will be close to 100%.
3. Clinical utility
3.1 (Differential) diagnostics: the tested person is clinically affected
(To be answered if in 1.9 “A” was marked)
3.1.1 Can a diagnosis be made other than through a genetic test?
No. | ☐ (continue with 3.1.4) | |
Yes, | ⊠ | |
Clinically | ⊠ | |
Imaging | ⊠ | |
Endoscopy | ☐ | |
Biochemistry | ☐ | |
Electrophysiology | ☐ | |
Other (please describe) |
3.1.2 Describe the burden of alternative diagnostic methods to the patient
PCG is a rare but severe blinding type of paediatric glaucoma. It is characterised by congenital elevation of intraocular pressure (IOP) resulting from significantly reduced aqueous outflow through a malfunctioning trabecular meshwork [1, 2]. The raised IOP results in progressive optic nerve damage and visual loss.
Children who are suspected of PCG will undergo repeated examinations under general anaesthesia, including corneal diameter measurements, IOP measurements, gonioscopy, optic nerve examination, refraction [30], ultrasound of the eye, and later on automated visual field testing can be introduced. Spectral domain optical coherence tomography can also be used to evaluate the optic nerve and retinal nerve fibre layer in PCG patients [31].
Genetic testing can help in the diagnosis and genetic counselling of families with PCG. If biallelic CYP1B1 or LTBP2 variants that affect function have been identified in the family, genetic testing of at-risk siblings as soon as possible after birth will avoid repeated examinations under anaesthesia in young children who do not carry the variants.
3.1.3 How is the cost effectiveness of alternative diagnostic methods to be judged?
PCG is responsible for 5% of childhood blindness [32] and accounts for up to 18% of children in blind institutions around the world [33]. Clinical diagnosis of young children with PCG can be challenging, especially for ophthalmologists in non-specialist centres. Patients will often require tertiary referral for accurate diagnosis and treatment. Genetic testing is important for the accurate diagnosis of children with an increased risk based on their family history. Optic nerve cupping may progress fast in newborns and can be reversible if normal IOP is restored rapidly [34].
3.1.4 Will disease management be influenced by the result of a genetic test?
No. | ☐ |
Yes. | ⊠ |
Therapy (please describe) | Biallelic CYP1B1 or LTBP2 variants that affect function confirm the diagnosis of PCG. There are medical and surgical treatments available in PCG and children can be treated at an early age to prevent visual loss |
Prognosis (please describe) | PCG is a potentially blinding disease and prognosis is largely related to the timing of presentation. Early diagnosis and prompt surgical treatment significantly improve the visual outcome |
Management (please describe) | PCG should be managed by specialists with expertise in this condition. Patients will require lifelong monitoring to ensure control of intraocular pressure. The management of paediatric glaucoma has improved considerably over the past few decades. Most cases are managed in tertiary referral centres and the mainstay of treatment is surgery 14. Supportive measures for those with sight impairment include involvement of social services, and regular refraction and correction to prevent amblyopia. Genetic counselling will also be offered to the family |
3.2 Predictive setting: The tested person is clinically unaffected but carries an increased risk based on family history
(To be answered if in 1.9 “B” was marked)
3.2.1 Will the result of a genetic test influence lifestyle and prevention?
If the test result is positive (please describe)
There are medical and surgical treatments available for PCG. If the test result is positive, the patient will be followed up regularly and appropriate treatment can be started early to prevent visual loss. Identification of an unaffected carrier will inform family planning.
If the test result is negative (please describe)
As there is only a 40% chance of picking up a genetic diagnosis, 60% will still have PCG but without a genetic cause. The result will help to inform family planning.
3.2.2 Which options in view of lifestyle and prevention does a person at-risk have if no genetic test has been done (please describe)?
The child will require regular outpatient clinic appointments and repeated examination under anaesthesia in early childhood followed by regular clinic visits to ensure he/ she is not developing signs of PCG. It can affect schooling and limit professions which require perfect vision. Hence, a clinically confirmed diagnosis can help in providing guidance regarding career choice.
3.3 Genetic risk assessment in family members of a diseased person
(To be answered if in 1.9 “C” was marked)
3.3.1 Does the result of a genetic test resolve the genetic situation in that family?
A molecular diagnosis in an affected individual can resolve the genetic situation in that family.
3.3.2 Can a genetic test in the index patient save genetic or other tests in family members?
If molecular testing has identified a CYP1B1 or LTBP2 or MYOC variant in the index patient, examination can identify and exclude disease in at-risk relatives. Most PCG cases (>80%) present within the first year of age [20, 21]. However, further genetic tests are required to determine the carrier status.
3.3.3 Does a positive genetic test result in the index patient enable a predictive test in a family member?
Yes
3.4 Prenatal diagnosis
(To be answered if in 1.9 “D” was marked)
3.4.1 Does a positive genetic test result in the index patient enable a prenatal diagnosis?
Yes
4. If applicable, further consequences of testing
Please assume that the result of a genetic test has no immediate medical consequences. Is there any evidence that a genetic test is nevertheless useful for the patient or his/her relatives? (Please describe)
Genetic testing for CYP1B1, LTBP2 and MYOC variants will provide a molecular diagnosis and help effective genetic counselling. However, acting upon the result in terms of termination is not normally advised as treatment can be provided. If biallelic variants that affect function have been identified in the family, genetic testing of at-risk siblings as soon as possible after birth will avoid repeated examinations under anaesthesia in young children who do not carry the variants.
References
Kupfer C, Kaiser-Kupfer MI. Observations on the development of the anterior chamber angle with reference to the pathogenesis of congenital glaucomas. Am J Ophthalmol. 1979;88(3 Pt 1):424–6.
Anderson DR. The development of the trabecular meshwork and its abnormality in primary infantile glaucoma. Trans Am Ophthalmol Soc. 1981;79:458–85.
Abu-Amero KK, Osman EA, Mousa A, et al. Screening of CYP1B1 and LTBP2 genes in Saudi families with primary congenital glaucoma: genotype-phenotype correlation. Mol Vis. 2011;17:2911–9.
Bejjani BA, Lewis RA, Tomey KF, et al. Mutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi Arabia. Am J Hum Genet. 1998;62:325–33.
Chakrabarti S, Ghanekar Y, Kaur K, et al. A polymorphism in the CYP1B1 promoter is functionally associated with primary congenital glaucoma. Hum Mol Genet. 2010;19:4083–90.
López-Garrido MP, Medina-Trillo C, Morales-Fernandez L, et al. Null CYP1B1 genotypes in primary congenital and nondominant juvenile glaucoma. Ophthalmology. 2013;120:716–23.
Lee JH, Ki CS, Kim HJ, et al. Analysis of copy number variation using whole genome exon-focused array CGH in Korean patients with primary congenital glaucoma. Mol Vis. 2011;17:3583–90.
Souzeau E, Hayes M, Ruddle JB, et al. CYP1B1 copy number variation is not a major contributor to primary congenital glaucoma. Mol Vis. 2015;21:160–4.
Ali M, McKibbin M, Booth A, et al. Null mutations in LTBP2 cause primary congenital glaucoma. Am J Hum Genet. 2009;84:664–71.
Narooie-Nejad M, Paylakhi SH, Shojaee S, et al. Loss of function mutations in the gene encoding latent transforming growth factor beta binding protein 2, LTBP2, cause primary congenital glaucoma. Hum Mol Genet. 2009;18:3969–77.
Azmanov DN, Dimitrova S, Florez L, et al. LTBP2 and CYP1B1 mutations and associated ocular phenotypes in the Roma/Gypsy founder population. Eur J Hum Genet. 2011;19:326–33.
Kaur K, Reddy AB, Mukhopadhyay A, et al. Myocilin gene implicated in primary congenital glaucoma. Clin Genet. 2005;67:335–40.
Akarsu AN, Tarucli ME, Aktan SG, et al. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum Mol Genet. 1996;5:1199–203.
Papadopoulos M, Cable N, Rahi J, Khaw PT, Eye BIG. Study Investigators. The British Infantile and Childhood Glaucoma (BIG) Eye Study. Invest Ophthalmol Vis Sci. 2007;48:4100–6.
Dandona L, Williams JD, Williams BC, Rao GN. Population-based assessment of childhood blindness in southern India. Arch Ophthalmol. 1998;116:545–6.
Alfadhli S, Behbehani A, Elshafey A, Abdelmoaty S, Al-Awadi S. Molecular and clinical evaluation of primary congenital glaucoma in Kuwait. Am J Ophthalmol. 2006;141:512–6.
Gencik A, Gencikova A, Ferák V. Population genetical aspects of primary congenital glaucoma. I. Incidence, prevalence, gene frequency, and age of onset. Hum Genet. 1982;61:193–7.
Gencik A. Epidemiology and genetics of primary congenital glaucoma in Slovakia. Description of a form of primary congenital glaucoma in gypsies with autosomal-recessive inheritance and complete penetrance. Dev Ophthalmol. 1989;16:76–115.
Elder MJ. Congenital glaucoma in the West Bank and Gaza Strip. Br J Ophthalmol. 1993;77:413–6.
Gramer E, Tausch M, Kraemer C. Time of diagnosis, reoperations and long-term results of goniotomy in the treatment of primary congenital glaucoma: a clinical study. Int Ophthalmol. 1996;20:117–23. 1997
Kolker AE, Hetherington JJ. Congenital glaucoma. Becker-Shaffer’s Diagnosis and Therapy of the Glaucomas. 5th edn. St. Louis: CV Mosby; 1983. p. 317–69.
McGinnity FG, Page AB, Bryars JH. Primary congenital glaucoma: twenty years experience. Ir J Med Sci. 1987;156:364–5.
Jay MR, Rice NSC. Genetic implications of congenital glaucoma. Metab Ophthalmol. 1978;2:257–8.
François J. Congenital glaucoma and its inheritance. Ophthalmologica. 1980;181:61–73.
Morin JD, Merin S, Sheppard RW. Primary congenital glaucoma: a survey. Can J Ophthalmol. 1974;9:17–28.
Hoskins DH, Kass MA. Becker-Shaffer’s diagnosis and therapy of the glaucoma. 6th edn. St Louis: CV Mosby. 1989;356.
Alanazi FF, Song JC, Mousa A, et al. Primary and secondary congenital glaucoma: baseline features from a registry at King Khaled Eye Specialist Hospital, Riyadh, Saudi Arabia. Am J Ophthalmol. 2013;155:882–9.
Fung DS, Roensch MA, Kooner KS, Cavanagh HD, Whitson JT. Epidemiology and characteristics of childhood glaucoma: results from the Dallas Glaucoma Registry. Clin Ophthalmol. 2013;7:1739–46.
Tamcelik N, Atalay E, Bolukbasi S, Capar O, Ozkok A. Demographic features of subjects with congenital glaucoma. Indian J Ophthalmol. 2014;62:565–9.
Faiq M, Sharma R, Dada R, Mohanty K, Saluja D, Dada T. Genetic, biochemical and clinical insights into primary congenital glaucoma. J Curr Glaucoma Pract. 2013;7:66–84.
Srinivasan S, Addepalli UK, Rao HL, Garudadri CS, Mandal AK. Spectral domain optical coherence tomography in children operated for primary congenital glaucoma. Br J Ophthalmol. 2014;98:162–5.
Gilbert CE, Rahi JS, Quinn GE. Visual impairment and blindness in children. In: Johnson GJ, Minassian DC, Weale RA, West SK, editors. The Epidemiology of Eye Disease. 2nd edn. London: Edward Arnold Ltd; 2003. p. 260–86.
Gilbert CE, Canovas R, Kocksch de Canovas R, Foster A. Causes of blindness and severe visual impairment in children in Chile. Dev Med Child Neurol. 1994;36:326–33.
Quigley HA. The pathogenesis of reversible cupping in congenital glaucoma. Am J Ophthalmol. 1977;84:358–70.
Acknowledgements
This work was supported by EuroGentest2 (Unit 2: “Genetic testing as part of health care”), a Coordination Action under FP7 (Grant Agreement Number 261469) and the European Society of Human Genetics. CY, PTK, and MM acknowledge the support of the National Institute for Health Research (NIHR) Biomedical Research Centre based at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, Moorfields Eye Charity and the Medical Research Council. MM acknowledges the support of the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Yu-Wai-Man, C., Arno, G., Brookes, J. et al. Primary congenital glaucoma including next-generation sequencing-based approaches: clinical utility gene card. Eur J Hum Genet 26, 1713–1718 (2018). https://doi.org/10.1038/s41431-018-0227-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0227-y