Abstract
Recurrent hydatidiform moles (RHM) are aberrant human pregnancies characterized by absence of, or abnormal, embryonic development, hydropic degeneration of chorionic villi, and hyperproliferation of the trophoblast. Biallelic mutations in two maternal-effect genes, NLRP7 and KHDC3L, underlie the causation of RHM in 60% of patients. We performed exome sequencing on a patient with six pregnancy losses, two miscarriages and four HM, and found no variants that affect the functions of the known genes. We found biallelic missense variants that affect conserved amino acids in PADI6 and segregate with the disease phenotype in the family. PADI6 is another maternal-effect gene and a member of the subcortical maternal complex that has been shown to have recessive variants that affect the gene function in four unrelated women with infertility who also experienced early embryonic arrest during preimplantation development after IVF. We demonstrated that PADI6 co-localizes with NLRP7 in human oocytes and preimplantation embryos and reviewed the morphology and genotypes of four products of conception from our patient. Our data expand the involvement of PADI6 to other forms of reproductive loss and highlight the commonality between infertility, miscarriages, and molar pregnancies, in some cases.
Similar content being viewed by others
Introduction
Hydatidiform mole (HM) is an aberrant human pregnancy characterized by abnormal embryonic development, hydropic chorionic villi, and excessive proliferation of the trophoblast. Most HMs are sporadic and not recurrent. They affect 1 in 600 pregnancies in western countries and up to 1 in 60 pregnancies in Southeastern Asia. Recurrent HM (RHM) are defined by the occurrence of at least two HM in the same patient and affects 1 to 9% of patients with a prior HM. To date, two maternal-effect genes, NLRP7 and KHDC3L, have been described to play causal roles in RHM and underlie the etiology in 55 and 5% of patients, respectively [1, 2]. The proteins coded by these two genes co-localize to the subcortical region of human oocytes and are absent from the cell-to-cell contact in early cleavage embryo stages [3]. The localization of NLRP7 and KHDC3L in human oocytes is similar to that of several other maternal-effect proteins in mice such as OOEP, NLRP2, NLRP5, TLE6, and PADI6. Some of these proteins were shown to interact when over-expressed in human cell lines [4] and under physiological conditions in mouse oocytes where they form a large multi-protein complex, called the subcortical maternal complex (SCMC), that is believed to contain additional interacting proteins [5].
In humans, beside the established roles of NLRP7 and KHDC3L in recurrent hydatidiform moles, variants in NLRP2 and NLRP5 have been found in one case of Beckwith–Wiedemann syndrome [6] and five patients with recurrent pregnancy loss and children with multilocus imprinting disturbance, respectively [7]. Recently, recessive variants that affect the functions of TLE6 [8] and PADI6 [9, 10] have been found responsible for primary female infertility and early development arrest during embryonic cleavage stages after assisted reproductive technologies.
To date, we have performed exome sequencing on 24 Chinese women with RHM and no variants that affect the functions of NLRP7 or KHDC3L were found. Here, we report the identification of a patient with two missense variants that affect conserved amino acids in PADI6 in a compound heterozygous state. We describe the re-evaluation of four of her conceptions and the co-localization of PADI6 with NLRP7 in human oocytes and early cleavage embryos.
Subjects and methods
The study was approved by the Chinese Committees for Ethical Review of Research involving Human Subjects at The First Affiliated Hospital (College of Medicine, Zhejiang University) and the McGill Institutional review Board (IRB# A01-M07–03 A). The patient and her relatives provided written consent to participate in the study. The patient is of Han Chinese origin and had a total of six pregnancy losses from natural spontaneous conceptions, two of them were initially diagnosed as miscarriages and four as HM. Beside her reproductive problem, the patient is in good health.
Genetic analyses
DNA was extracted from peripheral blood according to standard methods. Exome sequencing was performed at the McGill University and Genome Quebec Innovation Center (Montreal, Canada). The SureSelect Human All Exon Kit version 5 (Agilent Technologies, Santa Clara, CA, USA) was used for Exome enrichment. Exome library was then sequenced with 100-bp paired-end reads on the Illumina Hiseq2000 sequencer and the reads were aligned to the human reference genome (hg19). Variants nomenclature in PADI6 are given according to the following references, NM_207421.4 (cDNA) and NP_997304.3. Exon numbering is according to NG_032943.1. All variants detected have been submitted to the Leiden Open Variation Database (https://databases.lovd.nl/shared/genes/PADI6) (patient ID 00151816).
Characterization of the products of conception
Morphological evaluation and p57 immunohistochemistry was performed according to standard procedures as previously described [11]. Short tandem repeat (STR) genotyping was performed using an AmpFlSTR® Sinofiler™ PCR Amplification Kit (Sinofiler kit) (Applied Biosystems, Inc., Foster City, CA) as previously described [12].
Oocytes collection and confocal immunofluorescence
After obtaining written informed consent, spare oocytes and preimplantation embryos from patients undergoing in vitro fertilization or intra-cytoplamic sperm injection and preimplantation genetic diagnosis were collected at the McGill Reproductive Center. The oocytes and embryos were fixed with a microtubule-stabilizing buffer (0.1 M Pipes, pH 6.9, 5 mM MgCl2, 2.5 mM EGTA) containing 2.0% (v/v) paraformaldehyde, 0.5% (v/v) Triton X-100, 1 µM taxol, 10 units/ml aprotinin and 50% (v/v) deuterium oxide, washed and processed as previously described [3]. The following primary and secondary antibodies were used, Goat anti-NLRP7 (sc-50642, 1:100) (Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and Rabbit-anti-PADI6 (ab16480, 1: 100) (Abcam, Inc, CA, USA), Alexa 488-conjugated or Alexa 568-conjugated secondary antibodies (1:500) (Invitrogen, Thermo Fisher Scientific, Massachusetts, USA) were used. Fluorescence images were captured using Zeiss LSM780 Laser Scanning Confocal Microscope. Using the ZEN software (Zeiss provided software package), the Pearson’s correlation coefficient (PCC) was performed to quantify and check for co-localization.
Results and discussion
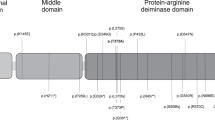
After whole exome sequencing, variant filtering was performed by filtering out variants with MAF > 1% in the following public databases ExAC (http://exac.broadinstitute.org/), 1000 Genome (http://www.internationalgenome.org/1000-genomes-browsers/), and EVS (http://evs.gs.washington.edu/EVS/). We only considered variants located in the coding regions (including missense, nonsense, frameshift, indels or canonical splice site changes) for downstream analysis. Finally, we selected for variants in homozygous or compound heterozygous states in genes that are not highly mutated in our in-house exomes. This led to seven candidate genes, of which five had missense variants with at least one variant predicted to affect the conserved amino acids by two prediction softwares, Polyphen and scaled CADD. Of these genes, only PADI6 was reported to play an important role in mouse and human infertility and early embryonic development arrest. The patient is compound heterozygous for two variants in PADI6, c.1793A > G, p.(Asn598Ser) and c.2045 G > A, p.(Arg682Gln). Variant c.1793A > G was not reported in any public databases (ExAC, 1000 Genome, EVS) while c.2045 G > A was found at a very low frequency (0.00000812) in ExAc. One of these variants, p.(Arg682Gln) was predicted to be disease-causing (polyphen = 1.0 and scaled CADD = 14) while the other, p.(Asn598Ser) was predicted to be benign (polyphen = 0.199, scaled CADD = 2.5), but both affect amino acids that are conserved in different mammalian species (Fig. 1). Sanger sequencing demonstrated the segregation of the two variants in the patient’s family, the inheritance of each variant from one parent, and the presence of only one of them in each of her unaffected sisters with normal pregnancies (Fig. 1).
Identification of two missense variants in PADI6. a Pedigrees structure of patient 587 and the segregation of the identified variants in her family. b Sanger sequencing confirmation of the two variants in the patient and the inheritance of each variant from one parent. c The conservation of the changed amino acid residues in different mammalian species. NM non-molar miscarriages, HM hydatidiform mole, POC product of conception. The POC are listed by their chronological order from left to right
Tissue sections from four available products of conception (POC) from the patient were available for re-evaluation and their morphological examination revealed hydropic and non-hydropic chorionic villi, as well as fetal vessels with nucleated red blood cells in all of them (Fig. 2). The 2nd, 5th, and 6th POC did not have trophoblastic proliferation and only the 4th POC had some chorionic villi with circumferential trophoblastic proliferation (Fig. 2b). Significant karyorrhexis was observed in some chorionic villi from the 4th POC (Fig. 2c). Based on these findings, the diagnoses of the 2nd, 5th, and 6th POC were concluded as non-molar miscarriages and only the 4th POC was diagnosed, by morphology, as a HM. This POC could not be classified into complete or partial because it does not have the typical features of neither type. p57KIP2 is the product of CDKN1C, a paternally imprinted, maternally expressed gene located on chromosome 11p15.5, and was used on the 2nd, 4th, and 5th POC. In androgenetic CHM which lacks maternal genome, p57KIP2 is not expressed in the nuclei of the cytotrophoblastic and stromal cells of the chrorionic villi. The 2nd and 5th POC were positive for p57 KIP2 expression in the nuclei of cytotrophoblastic and stroma cells. The 4th POC was also mostly positive, but had many negative nuclei in cytotrophoblastic and most villous stroma cells (Fig. 2d, e, f). STR genotyping was performed on one pregnancy, the 5th POC, along with parental DNA, and demonstrated its diploid biparental origin based on 14 markers on different chromosomes (Fig. 3a, b).
Examples of hematoxylin and eosin staining of sections from four products of conception (POC) from patient 587. a A microphotograph from the 2nd POC showing hydropic and non-hydropic chorionic villi with no trophoblastic proliferation. The inset shows a magnification of a fetal vessel. This POC was diagnosed as non-molar (NM) miscarriage. b A view from the 4th POC that was diagnosed as a HM showing a chorionic villous (indicated by CV) with circumferential trophoblastic proliferation and fetal vessels (one shown in the inset). Other chorionic villi from this HM showed significant karyorrhexis in (c). d, e, f p57 immunohistochemistry from this HM showing cytotrophoblastic cells with positive (arrows) and negative (arrowheads) nuclei. Clearly negative stromal nuclei are indicated by asterisks on their left. Note in (e, f) all stromal nuclei are negative. g A microphotograph of the 5th POC diagnosed as non-molar miscarriage showing hydropic and non hydropic chorionic villi, with fetal vessels. h A microphotograph from the 6th POC diagnosed as non-molar miscarriage showing several chorionic villi with no trophoblastic proliferation
STR genotyping of one product of conception (POC) from patient 587 and co-localization of PADI6 and NLRP7 proteins. a Allelic inheritance at three markers showing the transmission of one parental allele at each. For example, in the POC, at marker D19S433, allele 15.2 is inherited from the patient and allele 15 from her partner. b Alleles at all amplifiable markers. c Co-localization of PADI6 (red) and NLRP7 (green) in a GV oocyte (top panel) and a hatching blastocyst (bottom panel) with Pearson correlation coefficient scores 0.81 and 0.87, respectively, by confocal microscopy
PADI6 codes for peptidylarginine deiminase, a member of a class of Ca2+-dependent enzyme that posttranslationally modifies arginine residues within peptides (peptidylarginine) to citrulline. This process, called citrullination, plays important roles in the formation of rigid structures such as hair, skin, and myelin sheath. Padi6 knockout in mice leads to infertility that affects only null females [13]. Oocytes from Padi6−/− lack a specialized cytoskeletal structure called cytoplasmic lattices (CPL) and consequently have abnormal organelle positioning and redistribution, reduced amounts of ribosomal and messenger RNA, altered de novo protein synthesis, and impaired embryonic genome activation [13,14,15]. In mice, PADI6 co-localizes with KHDC3 (the ortholog of human KHDC3L) and NLRP5 and is part of the SCMC. Because (i) PADI6 is a member of the mouse maternal subcortical complex, (ii) plays a causal role in early embryonic development in humans, and (iii) is mutated in our patient with recurrent miscarriages and one HM, we investigate its localization relative to NLRP7 in human oocytes and early cleavage embryos. The results of this analysis demonstrated that PADI6 in human oocytes has a similar localization to that observed in mice and is abundant in the subcortical region and co-localizes with NLRP7 (Pearson coefficient = 0.81) (Fig. 3). Similar co-localization was also observed at the morula and blastocyst stages (Fig. 3).
In humans, functional PADI6 variants have been reported in one familial case (consisting of three sisters) and 3 other unrelated patients with primary infertility and early development arrest during embryonic cleavage stages after in vitro fertilization [9, 10]. Hydatidiform mole is another form of early arrested embryonic development, but the pregnancy is retained by the mother to 8–12 weeks. Among reported patients with biallelic functional PADI6 variants, our patient is the first to have detectable pregnancies. One possible explanation of the difference between the reproductive outcomes of our and previously reported patients is the fact that she has two missense variants while the others, each, had one or two protein-truncating variants. The missense variants in our patient may have a milder effect on the protein than protein-truncating variants, and consequently may delay embryonic arrest by few days and be permissive for blastocyst implantation and differentiation of some embryonic tissues. Similar observations were made in patients with RHM and biallelic functional NLRP7 variants where missenses, but not protein-truncating variants, were associated with the presence of some embryonic tissues [11]. Furthermore, the association between diploid biparental HM, miscarriages, and infertility has also been observed in many patients with biallelic functional variants in NLRP7 and some of their HM were diagnosed by morphology as non-molar miscarriages, partial HM (because of their mild trophoblastic proliferation), non-classical HM [16], or not easy to classify HM [17]. Definitely, infertility appears to be more common among patients with biallelic functional variants in PADI6, but has also been observed in five (out of 62) unrelated cases with recessive functional variants in NLRP7 [18, 19] (and unreported observations). In these five cases, the sisters of the probands with RHM had infertility of unexplained clinical origin and biallelic NLRP7 variants that affect the function.
While it is well-known that the primary defect in patients with RHM and biallelic functional variants in NLRP7 is in their oocytes, the only documentation of how their embryos develop during early stages has been shown in one patient by Sills et al. [20]. In this patient, 15 oocytes were retrieved and fertilized, but the resulting embryos were not of good quality and all arrested between day 3 and 6 like those from patients with recessive functional PADI6 variants [10]. In conclusion, the data described in this report implicate PADI6 in miscarriages and molar pregnancies and highlight the commonality between defects in NLRP7 and PADI6, in their localization in human oocytes and early embryos, the development of their embryos during early cleave stages, and their association with HM, miscarriages, and infertility albeit some differences.
References
Murdoch S, Djuric U, Mazhar B, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet. 2006;38:300–2.
Parry DA, Logan CV, Hayward BE, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet. 2011;89:451–8.
Akoury E, Zhang L, Ao A, Slim R. NLRP7 and KHDC3L, the two maternal-effect proteins responsible for recurrent hydatidiform moles, co-localize to the oocyte cytoskeleton. Human Reprod. 2015;30:159–69.
Zhu K, Yan L, Zhang X, et al. Identification of a human subcortical maternal complex. Mol Hum Reprod. 2015;21:320–9.
Li L, Baibakov B, Dean J. A subcortical maternal complex essential for preimplantation mouse embryogenesis. Dev Cell. 2008;15:416–25.
Meyer E, Lim D, Pasha S, et al. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome). PLoS Genet. 2009;5:e1000423.
Docherty LE, Rezwan FI, Poole RL, et al. Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun. 2015;6:8086.
Alazami AM, Awad SM, Coskun S, et al. TLE6 mutation causes the earliest known human embryonic lethality. Genome Biol. 2015;16:240.
Maddirevula S, Coskun S, Awartani K, Alsaif H, Abdulwahab FM, Alkuraya FS. The human knockout phenotype of PADI6 is female sterility caused by cleavage failure of their fertilized eggs. Clin Genet. 2017;91:344–5.
Xu Y, Shi Y, Fu J, et al. Mutations in PADI6 cause female infertility characterized by early embryonic arrest. Am J Hum Genet. 2016;99:744–52.
Nguyen NM, Zhang L, Reddy R, et al. Comprehensive genotype-phenotype correlations between NLRP7 mutations and the balance between embryonic tissue differentiation and trophoblastic proliferation. J Med Genet. 2014;51:623–34.
Zheng XZ, Hui P, Chang B, et al. STR DNA genotyping of hydatidiform moles in South China. Int J Clin Exp Pathol. 2014;7:4704–19.
Esposito G, Vitale AM, Leijten FP, et al. Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol. 2007;273:25–31.
Lu X, Gao Z, Qin D, Li L. A maternal functional module in the mammalian oocyte-to-embryo transition. Trends Mol Med. 2017;23:1014–23.
Yurttas P, Vitale AM, Fitzhenry RJ, et al. Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development. 2008;135:2627–36.
Sebire NJ, Savage PM, Seckl MJ, Fisher RA. Histopathological features of biparental complete hydatidiform moles in women with NLRP7 mutations. Placenta. 2013;34:50–6.
Brown L, Mount S, Reddy R, et al. Recurrent pregnancy loss in a woman with NLRP7 mutation: not all molar pregnancies can be easily classified as either “partial” or “complete” hydatidiform moles. Int J Gynecol Pathol. 2013;32:399–405.
Qian J, Deveault C, Bagga R, Xie X, Slim R. Women heterozygous for NALP7/NLRP7 mutations are at risk for reproductive wastage: report of two novel mutations. Hum Mutat. 2007;28:741.
Slim R, Bagga R, Chebaro W, Srinivasan R, Agarwal N. A strong founder effect for two NLRP7 mutations in the Indian population: an intriguing observation. Clin Genet. 2009;76:292–5.
Sills ES, Obregon-Tito AJ, Gao H, et al. Pathogenic variant in NLRP7 (19q13.42) associated with recurrent gestational trophoblastic disease: data from early embryo development observed during in vitro fertilization. Clin Exp Reprod Med. 2017;44:40–6.
Acknowledgements
This work was supported by the Canadian Institute of Health Research [MOP-130364 and CCI-125687 to RS] and by the National Natural Science Foundation of China [81261120569 and 81370729 to JHQ].
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Qian, J., Nguyen, N.M.P., Rezaei, M. et al. Biallelic PADI6 variants linking infertility, miscarriages, and hydatidiform moles. Eur J Hum Genet 26, 1007–1013 (2018). https://doi.org/10.1038/s41431-018-0141-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-018-0141-3
This article is cited by
-
Imprinting disorders
Nature Reviews Disease Primers (2023)
-
Loss of the Maternal Effect Gene Nlrp2 Alters the Transcriptome of Ovulated Mouse Oocytes and Impacts Expression of Histone Demethylase KDM1B
Reproductive Sciences (2023)
-
A novel heterozygous variant in PANX1 causes primary infertility due to oocyte death
Journal of Assisted Reproduction and Genetics (2023)
-
A novel homozygous mutation in the PADI6 gene causes early embryo arrest
Reproductive Health (2022)
-
Novel biallelic mutations in PADI6 in patients with early embryonic arrest
Journal of Human Genetics (2022)