Abstract
Myxobacterial predation on bacteria has been investigated for several decades. However, their predation on fungi has received less attention. Here, we show that a novel outer membrane β-1,6-glucanase GluM from Corallococcus sp. strain EGB is essential for initial sensing and efficient decomposition of fungi during predation. GluM belongs to an unstudied family of outer membrane β-barrel proteins with potent specific activity up to 24,000 U/mg, whose homologs extensively exist in myxobacteria. GluM was able to digest fungal cell walls efficiently and restrict Magnaporthe oryzae infection of rice plants. Genetic complementation with gluM restored the fungal predation ability of Myxococcus xanthus CL1001, which was abolished by the disruption of gluM homolog oar. The inability to prey on fungi with cell walls that lack β-1,6-glucans indicates that β-1,6-glucans are targeted by GluM. Our results demonstrate that GluM confers myxobacteria with the ability to feed on fungi, and provide new insights for understanding predator-prey interactions. Considering the attack mode of GluM, we suggest that β-1,6-glucan is a promising target for the development of novel broad-spectrum antifungal agents.
Similar content being viewed by others
Introduction
Myxobacteria are ubiquitous Gram-negative soil bacteria with a complex developmental life cycle and the capacity to influence the structure of ecological communities [1]. They can be classified as proteolytic, cellulolytic and chitinolytic according to the organic matter on which they feed [2]. Proteolytic myxobacteria are associated with feeding on a broad range of soil bacteria and fungi [2]. The cytotoxic mechanism that mediates the killing of bacterial prey has been investigated primarily in the myxobacterial model strain Myxococcus xanthus DK1622 [3,4,5]. However, ecological and biochemical factors that are involved in the myxobacterial predation of fungi are poorly understood.
The fungal cell wall is a highly complex structure that constitutes a shield protecting the cell from environmental stresses [6]. Therefore, it is the initial barrier that myxobacteria must overcome in order to prey on fungi. Cell wall degrading enzymes (CWDEs) play important roles in the decomposition of fungal cell walls by plants and biological control agents [7, 8]. However, myxobacterial CWDEs have not been investigated for their function in fungal cell wall lysis. So far, studies of antifungal CWDEs are limited to chitinases and β-1,3-glucanases [9]. This is unsurprising because since chitin and β-1,3-glucans are the predominant structural components of fungal cell walls [6]. In contrast, β-1,6-glucan is a comparatively minor component that crosslinks cell wall proteins to the β-1,3-glucan/chitin layer. Deletion of genes related to β-1,6-glucan synthesis and inhibition of the corresponding biosynthetic enzymes were found to be lethal for Candida albicans, Cryptococcus neoformans, and Colletotrichum graminicola, indicating that β-1,6-glucan is essential for the construction of rigid fungal cell walls [10,11,12]. Accordingly, β-1,6-glucanases may contribute to the efficient disorganization and further degradation of fungal cell walls during predation. Indeed, knockout of β-1,6-glucanase Tvbgn3 in Trichoderma virens decreased its biocontrol efficiency [13]. However, the antifungal properties of β-1,6-glucanases are largely unknown.
Outer-membrane vesicles (OMVs) derived from the cell envelope of Gram-negative bacteria play important roles in diverse bacterial functions [14]. Notably, OMVs of M. xanthus are directly involved in the lysis of prey cells [4]. A large number of proteins have been detected in these OMVs, a subset of which are specifically located or enriched in the vesicles [15]. Numerous β-barrel outer membrane proteins are found to be involved in a broad repertoire of cell functions [16]. Among these outer membrane proteins, a few are membrane-intrinsic β-barrel enzymes. Such membrane-intrinsic β-barrel enzymes can function as sentinels that respond to perturbations in the outer membrane permeability barrier [17]. However, the specific functions of M. xanthus OMV proteins remain largely unknown.
Fungi are devastating plant pathogens, causing enormous economic losses to global crop production [18]. Biocontrol agents have been promoted as replacements to synthetic pesticides due to the adverse effects associated with the excessive use of chemicals in agriculture [19]. Although myxobacteria have the potential to be used as biological control agents [20], their modes of action are poorly understood on a molecular level. We recently evaluated Corallococcus sp. strain EGB as a promising biocontrol agent [21] and hypothesized that intrinsic CWDEs are crucial in the predation process. Here, we show that a novel OM β-1,6-glucanase GluM, belonging to a yet unknown glycoside hydrolase family, is a key factor in the predation of the rice pathogen Magnaporthe oryzae. Our results provide a new potential avenue for the biocontrol of fungal phytopathogens.
Materials and methods
Culture conditions
Corallococcus sp. strain EGB (China Center for Type Culture Collection, CCTCC M2012528) was cultured on VY/4 plates (0.1% yeast cells (Angel, China) and 0.1% CaCl2, pH 7.0, w/v) at 30 °C; liquid VY/4 medium was used to prepare the supernatant liquid enzyme (SUP). M. xanthus DK1622 was cultured on CYE [22] or VY/4 at 28 °C. Malassezia restricta (CBS 7877) and Neurospora crassa (Agricultural Culture Collection of China, ACCC 32256) were cultured on modified Dixon’s medium [23] or VY/4 at 28 °C. M. oryzae Guy11 was used as a model of plant-pathogenic fungus in this study. The conditions for growth and conidiation were the same as published earlier [24]. Complete medium (CM) or PDA medium was used for the vegetative growth of strain Guy11 when necessary [25]. For conidiation, strain Guy11 was incubated on SDC medium (100 g of straw, 40 g of corn powder, and 15 g of agar in 1 L of distilled water) at 28 °C in the dark for 7 days and constant illumination for 3 days [26]. Conidia harvested from cultures were resuspended to 5 × 104 conidia/mL in a 0.2% gelatin solution. All the strains, plasmids and primers used in this study are listed in Tables S1–3.
Bio-control assay
To determine the antifungal activity of strain EGB against filamentous fungi (M. oryzae and N. crassa), prey strains were cultured on VY/4 medium for 2–3 days [21], after which the myxobacteria were inoculated around the fungal colonies. Additionally, a membrane-separated co-incubation of the strains EGB and M. oryzae Guy11 was carried out to test if direct cell contact is required for the antifungal effect (molecular cut-off 10 kDa). For predation assays using M. restricta, the fungus was first cultured on modified Dixon’s medium for about 3–4 days, and strain EGB was then inoculated on top of the fungal colonies. The growth of fungi and the dispersed colony of myxobacteria were observed 2–4 days later. The cellular morphology and cell wall integrity were observed with the use of a light microscopy when necessary.
To determine the antifungal activity of the extracellular secretion, the supernatant of the culture (SUP) was prepared as described previously [21]. The supernatant was inactivated at 100 °C for 5 min and the crude extractions of low molecular weight substances (SUF) were prepared by ultrafiltration (molecular cut-off 10 kDa), which were used as a control. For the rice seedling spraying assay, two-week-old seedlings of rice (Oryza sativa cv. CO39) were sprayed with 5 mL conidial suspension (5 × 104 spores/mL) for each treatment when necessary. The treatments were designed as follows: (1) rice seedlings spray-incubated with strain Guy11; (2) rice seedlings sprayed with strain Guy11 mixed with 5 mL SUF; (3) 5 mL SUP sprayed onto rice seedlings 1 day after the strain Guy11 spray treatment (SUP, 1 day later); (4) rice seedlings spray-incubated with strain Guy11 1 day after spray-treatment with 5 mL SUP (SUP, 1 day before); (5) rice seedlings sprayed with strain Guy11 mixed with 5 mL SUP (SUPM); (6) Control setup of rice seedlings prepared without any treatment. The rice plants were kept in a growth chamber at 25 °C with 90% humidity and in the dark for the first 24 h, followed by a 12 h/12 h light/dark cycle. The lesions from the leaves were photographed and scored. The rice blast scores including the control efficiency and disease index were analyzed according to 0–5 scores ranking system [27]. Five landmark lesion types were established for scoring purpose: 0, no lesion; 1, pinhead-sized brown specks; 2, 1.5 mm brown spots; 3, 2–3 mm grey spots with brown margins; 4, many elliptical grey spots longer than 3 mm; 5, coalesced lesions infecting 50% or more of the leaf area. The quantification of lesions from the rice leaves was then performed with types 1–5. The index of disease severity was calculated as follows: disease index = (5 N5 + 4N4 + 3N3 + 2N2 + 1N1 + 0N0)/5N × 100; where N0 − N5 is the number of leaves in each degree (0–5) and N is the number of total leaves investigated [28]. Control efficacy was calculated as (disease index of the control−disease index of the treatment)/disease index of the control × 100%.
Quantitative PCR analysis
Dispersed cells of strain EGB cannot be prepared due to the cell-cell adhesion. Hence, to quantitatively calculate the cells of strain EGB in fungal predation experiments, the spore suspension (5 × 104 spores/mL) without any vegetative cells was prepared (Methods in supplementary materials). For predation assay, the treatments were designed as follows: 10 µL strain EGB or Guy11 (5 × 104 spores/mL) were placed on VY/4 plate individually, and 10 µL strains EGB and Guy11 were mixed together and then placed on VY/4 plate at 28 °C for about 4 days. For qPCR, the agar samples were ground into fine powder by liquid nitrogen refrigeration, and the total DNA was isolated from the samples using a beating method (FastDNA SPIN Kit, MP Biomedicals, USA) following the manufacturer’s instructions. The qPCR was performed with three independent samples in three sets of experimental replicates (Methods in supplementary materials). The relative bacterial and fungal biomasses were quantified from the copies of strain EGB 16S rRNA or strain Guy11 28S rRNA genes, and the growths of strains EGB or Guy11 in a separate culture were tested as controls.
β-1,6-glucanase activity assay
The β-1,6-glucanase activity assay was performed using yeast glucan as substrate, quantifying the amount of released reducing sugar by using the dinitrosalicylic (DNS) acid method as described previously [29]. A mixture comprising 1 mL of 0.5% yeast glucan in 50 mM Tris-HCl buffer (pH 7.0), supplied with 10 µL of appropriately diluted enzyme, was incubated at 50 °C for 10 min. The absorbance at 540 nm (OD540) was determined photometrically using a series of glucose solutions for sample quantification. One unit of β-1,6-glucanase activity is defined as the amount of enzyme that releases 1 µmol of reducing sugar per minute under the described assay conditions.
Purification, identification and characterization of β-1,6-glucanase from Corallococcus sp. strain EGB
The supernatant of a culture of strain EGB was prepared as described previously [21]. For purification of the β-1,6-glucanase, the supernatant was fractionated at 40–80% saturation with ammonium sulfate, followed by centrifugation at 15,000 g for 20 min, then the pellet dissolved in a volume of 50 mM Tris-HCl (pH 7.0). The β-1,6-glucanase GluM was purified from the supernatant using the Sephadex G-75 (GE Healthcare) as an affinity matrix. Briefly, the dissolved solution was mixed with Sephadex G-75 at 4 °C for 4 h with continuous stirring, after which the resin was washed by decanting with 50 mM Tris-HCl buffer (pH 7.0). The β-1,6-glucanase was then released from the resin by adding yeast glucan. The released enzyme was the recovered by centrifugation at 12,000 g for 20 min and the resulting supernatant was subject to ammonium sulfate precipitaton (80% saturation). After centrifugation (15,000 g for 20 min), the resulting pellet was suspended in a minimum volume of 50 mM Tris-HCl buffer (pH 7.0).
The purity of the β-1,6-glucanase GluM preparations was analyzed by HPLC analysis (Shodex Protein KW802.5 column, 8 × 300 mm, 5 μm, Showa Denko America Inc., New York, NY, UV absorbance 280 nm) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gel was cut into two strips, of which the first sample strip was used for Coomassie brilliant blue R250 staining and the second was used for zymogram analysis after renaturation as described previously [30]. Briefly, the first step of the protein renaturation was the removal of SDS from the gel strip with two 30 min incubations in a 2.5% (v/v) Triton X-100 solution, followed by two consecutive incubations in 100 mM Tris-HCl buffer (pH 8.0). The gel was then soaked in 50 mM Tris-HCl buffer (pH 7.0), followed by placing the gel onto the agar plate containing 0.5% yeast glucan for 6 h at 37 °C. The stained gel band corresponding to the location of the transparent zone from the zymogram analysis was excised and analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) peptide mass fingerprinting (PMF), and the results of PMF were interpreted by referencing the Mascot database [31].
The substrate specificity of the purified β-1,6-glucanase GluM was determined with various substrates, including yeast glucan (Angel, China), pustulan (Elicityl-OligoTech, France), laminarin, mannan, cellulose (Sigma-Aldrich, USA), salecan and BIWP2 [32, 33]. General properties of GluM, including optimal pH and temperature, were investigated using yeast glucan as the substrate. For determination of the hydrolysis pattern, GluM was incubated with 10 mg/mL yeast glucan, laminarin, pustulan and extracted ASDN faction (Methods in supplementary materials) at 30 °C for 10 h, after which the soluble carbohydrate composition was analyzed by TLC on silica gel 60 plates (Merck, Germany) using n-butanol-methanol-H2O (8:4:3, v/v/v) as the solvent system [34]. The reaction products were visualized by spraying a sulfuric acid-methanol (1:1, v/v) solution onto the plate, followed by baking at 95 °C for 10 min. The distribution of hydrolyzed polysaccharides was analyzed by HPLC using a Cosmosil Sugar-D column (KS-805, 8 mm by 300 mm; Nacalai Tesque Co., Japan) that was maintained at 50 °C and a RID detector. The mobile phase used in the system consisted of ultrapure water at a flow rate of 0.8 mL/min.
In the modified pull-down assay, the purified β-1,6-glucanase was incubated for 4 h at 4 °C with the insoluble glucans including yeast glucan (Angel, China), pachyman (Sigma-Aldrich, USA) and dextran (Sephadex G-25, GE Healthcare) with glucose as the constituent unit, and chitosan (Sigma, China) as a non-glucosidic control polysaccharide. Alternatively, the supernatant of strain EGB (SUP) was also incubated with the insoluble crude cell walls of strain Guy11 (Methods in supplementary materials) at 4 °C for 4 h. The amount of GluM remaining in the supernatant, co-precipitating with the substrate and from the crude cell walls, was examined by SDS-PAGE and tested for β-1,6-glucanase activity as described above.
Assays for conidia germination, vegetative growth, and pathogenicity
To determine the effects of the β-1,6-glucanase GluM on the conidia germination, the conidia were incubated on Gel Bond film (FMC Bioproducts, Rockland, ME, USA) at 28 °C in the presence of purified GluM fractions derived from the supernatant of strain EGB. The thermally inactivated SUP (TSUP) and SUF (SUP ultrafiltrate; <10 kDa) were used as control. After 4 and 8 h of incubation, the germination tubes and appressoria were observed using differential interference contrast microscope (DIC, Zeiss Axio Observer A1). For identification of effects on the cell walls of conidia and mycelia, the prepared conidia and mycelia were treated with purified GluM, and observed by scanning electron microscope (SEM, Hitachi SU8010) and transmission electron microscopy (TEM, Hitachi HT7700). Additionally, the effects of GluM treatment on yeast cell walls were also analyzed by TEM.
For the sheath infection assay, conidia were injected into leaf sheaths of rice seedlings (O. sativa cv. CO-39) with a 1 mL syringe. After 24 or 48 h of incubation at room temperature under humid conditions (90%), the inner epidermis of infected sheaths was observed under a microscope, and the penetration and infectious hypha expansion in rice tissue were classified into four types as described previously [26].
Assays for appressorium formation
Conidial suspensions were placed on cover glass (Fisher-brand, UK) and incubated at 28 °C for 8 h to induce appressorium formation, after which the water surrounding the appressoria was replaced with an equal volume of the purified GluM solution, followed by incubation at 28 °C for 4 h, after which the solution was replaced with an equal volume of 2–4 M glycerol. Morphological observation of the appressoria was performed followed the treatments.
Assays for the cell viability
The cells of M. restricta and conidia of strain Guy11 were prepared as described above, and the spores of N. crassa were prepared on VY/4 at 28 °C after 4–5 days growth. The cells or conidia suspension with a concentration of 1 × 104 /mL was mixed with GluM (3 U/mL), followed by incubation at 30 °C for various reaction times. The cells or conidia without any treatment were used as controls. Cell viability was tested via continuous dilution on PDA or Dixon’s medium followed by counting of fungal colonies after incubation. The numbers of fungal colonies were regarded as 100% in the control experiment and the relative cell survival rates from different treatments were calculated accordingly.
The effect of GluM toward mycelia was also evaluated. To do so, mycelia of strain Guy11 were collected and washed with Tris-HCl buffer, mixed with GluM (3 U/mL), and then incubated at 30 °C for various reaction times. The mycelia were transferred onto a PDA plate and incubated at 28 °C, and the colony diameter was recorded after 5 days. Thermally inactivated GluM was used as control.
Predation test of M. xanthus strains
M. xanthus DK1622 and its derivative strains CL 1001 and CL 1002 were used in the predation test. Strain Guy11 was cultured on VY/4 medium at 28 °C for 2–3 days until the colony diameters reached approximately 2 cm, after which strains DK1622, CL1001 and CL1002 were inoculated around the fungal colonies. The growth of the fungal hyphae at the contact zone with the myxobacterial strains was observed by light microscopy (Olympus CX31). To examine the predatory behavior of strains EGB, DK1622, CL1001 and CL1002, the cells were collected from cultures and washed twice with TPM buffer, followed by adjusting the density to 5 × 108 cells/mL. An aliquot comprising 3 µL of the myxobacterial cell suspension was co-cultured with strain Guy11 on TPM plates. The movement of the strains was recorded after 3 days of cultivation. A detailed observation of the myxobacterial colonies was achieved by continuous video recording over a time period of 6 h.
Statistical analysis
All experiments were designed and performed in triplicate, and mean values were reported with standard deviations. Error bars denote st. dev. (SD), and values with different letters indicate statistically significant differences (p < 0.01), which were statistically evaluated using SPSS software (ver. 22.0; IBM Corp., Armonk, NY, USA). ANOVA was used for comparison of two specific treatments, and a t-test was used to establish significance.
Results
Corallococcus sp. strain EGB can prey on M. oryzae Guy11 and control rice blast disease
M. oryzae is a model plant pathogen for the study of plant–microbe interactions [35]. The biocontrol effect of strain EGB against rice blast caused by the plant pathogen M. oryzae was evaluated. Strain EGB was able to feed on M. oryzae Guy11 in the plate assay, resulting in dilapidated fungal colonies surrounding by the growing cells of strain EGB. The growth of strain Guy11 was not affected when strains EGB and Guy11 were separated by a semi-permeable membrane (Fig. 1a). This result suggests that the role of low molecular weight secondary metabolites is negligible in the biocontrol of M. oryzae by strain EGB. To identify the viability of strain Guy11 from the dilapidated colony, agar blocks P1, P2 and P3 were transferred to fresh PDA or VY/4 plates. Typical growth of strain EGB (P1 and P2) was observed on VY/4 plates, whereas growth of strain Guy11 on PDA plates was reduced (P2) as compared to controls (P3) (Figs. S1a, b). Strain Guy11 (P2) could not grow on VY/4 plates because of the growth of strain EGB. Considering the formation of filamentous aerial hyphae, the viability of M. oryzae Guy11 cannot be accurately determined using colony-forming units (CFU). Hence, qPCR analysis was performed, and the results showed that the relative biomass of strain Guy11 decreased by 83% from the predation of strain EGB, whereas the relative biomass of strain EGB increased by 250% (Fig. 1b, Fig. S2). These results indicate that the growth of strain EGB increased from prey-killing when strains EGB and Guy11 are co-cultivated. Cell-free culture supernatants of strain EGB (SUP) were then tested for their potential to control rice blast disease. SUP exhibited prominent biocontrol efficacy in the rice seedling infection assays. A significant reduction in number of lesions with different spot sizes was observed 7 days post-inoculation (Fig. 1c, Fig. S1d, e). We found that SUP showed protective and curative effects against strain Guy11, with the best biocontrol efficiency (84%) and healthy plant growth observed when SUP was sprayed as a mixture with conidia of strain Guy11 (SUPM) (Fig. 1d, Fig. S1c). Otherwise, small molecules in SUF (SUP ultrafiltrate; <10 kDa) exhibited no obvious effect on strain Guy11 pathogenicity (Fig. 1c, Fig. S1e), and it was similar to what was observed in the plate membrane separation assay (Fig. 1a).
Biocontrol of M. oryzae using Corallococcus sp. strain EGB. a Co-cultured strains Guy11 and EGB on VY/4 plates with (Guy11//EGB) or without (Guy11 + EGB) membrane separation. Solid arrow indicates a monocultural colony of strain Guy11, dashed arrow indicates the collapsed colony of strain Guy11, and dotted line indicates the semi-permeable membrane (molecular cut-off 10 kDa). b Relative biomass analysis of strains EGB and Guy11 from the plates by quantitative PCR. Strain EGB or Guy11 was cultivated separately or co-cultured on VY/4 plates. Genomic DNAs on the cultivated plates were isolated for quantitation of relative biomass of strain EGB or Guy11. c Effects of strain Guy11 infection in conjunction with SUP and SUF treatments. d Disease index and biocontrol efficiency. All rice seedlings were treated with strain Guy11 except for the control sample. Guy11: M. oryzae; EGB: strain EGB; SUP: cell-free culture supernatants of strain EGB; SUF: SUP ultrafiltrate (<10 kDa); SUP (1 day later) and SUP (1 day before): SUP was sprayed onto rice seedlings 1 day after or 1 day before the strain Guy11 spray treatments, respectively; SUPM: rice seedlings sprayed with strain Guy11 mixed with SUP. Data show the means of three biological replicates. Error bars denote ± standard error of the mean st. dev. (SD), and values with different letters (a–d) indicate statistically significant differences from different treatments (p < 0.01)
Purification and characterization of an outer membrane β-1,6-glucanase from strain EGB
According to the results described above and our previous observations [21], it was hypothesized that a glucan hydrolase from strain EGB may play a key role in the control of fungal infections. Using Sephadex G-75 affinity purification, a hydrolase designated as GluM (MH747076) was isolated from the culture supernatant of strain EGB with high purity (>92%) (Fig. S3). Based on the substrate specificity of GluM, the enzyme was classified as a β-1,6-glucan hydrolase (Fig. 2a, Table S4). The protein sequence of GluM was identified by peptide mass fingerprinting using tryptic peptide fragments of the purified enzyme, which was functionally verified by recombinant expression in E. coli BL21(DE3) (Tables S5 and 6). Sequence analysis did not reveal any characteristic functional domains in the β-1,6-glucanase GluM except for a putative carboxypeptidase regulatory domain and a TonB-dependent receptor domain (Fig. S4a). However, GluM contains characteristic transmembrane β-strand domains (Figs. S4b, c), a typical feature of outer membrane proteins. Western blot analysis also indicated that GluM was located in the membrane fraction and outer membrane vesicles (Figs. S4d, e). Although GluM did not share identity with known glycoside hydrolases, it shared 40–80% identity with TonB receptor proteins from myxobacteria, and was highly identical (73% identity with 100% query coverage) with the with the outer membrane protein Oar (OmpA-related protein, AAB27614) from M. xanthus DK1622 (Fig. S5 and Table S7). Therefore, GluM is the first representative of a yet unknown glycoside hydrolase (GH) family.
Purification of β-1,6-glucanase GluM and its products and substrates binding analysis. a SDS-PAGE analysis of the purified GluM (GluM-S) from strain EGB (lane 1) and recombinant GluM (GluM-B) from E. coli BL21 (DE3) (lane 2), and transparency caused by GluM-S hydrolysis of yeast glucan as revealed by zymogram analysis (lane 3, indicated by the arrow). b, d TLC and HPLC analysis of the released carbohydrates from GluM hydrolysis reactions (10 U/mL, 40 °C for 10 h) with yeast glucan (water insoluble), laminarin and pustulan (water soluble) (1%, w/v) as the substrates. G1 to G5 refer to the standard maltooligosaccharides glucose, maltose, maltotriose, maltotetraose and maltopentaose. c SDS-PAGE analysis of the adsorbed GluM from the pull-down assay with chitosan, yeast glucan, pachyman and dextran as substrates (2%, w/v). The protein concentration of GluM used in these assays was 60 µg/mL
The enzymatic activity studies showed that GluM was most active at 50 °C and pH 7.0, showing a specific activity of up to 24,000 U/mg using yeast glucan as a substrate (Fig. S6). Furthermore, the enzyme exhibited strong hydrolysis activities toward BIWP2, and to a lesser extent toward laminarin and pustulan (Fig. 2b, d, Table S4). Nevertheless, their molecular weight distribution of the tested substrates was remarkably changed after GluM treatment (Fig. 2d), indicating that GluM is able to alter the structural integrity of the tested substrates by the specific hydrolysis of β-1,6 glucosidic linkages.
Sephadex G-75 was successfully used as an affinity matrix during purification of the β-1,6-glucanase. We consequently deduced that a carbohydrate-binding module (CBM) may exist in GluM. To verify this assumption, the binding of GluM to insoluble polysaccharides was tested. Our results showed that GluM bound to yeast glucan, pachyman, dextran and the cell wall fraction of strain Guy11, but was not able to bind chitosan (Fig. 2c, Fig. S7). We therefore concluded that GluM is able to bind to glucose-based polymers to various extents, but not polymers composed of other sugars. The binding ability of GluM to the cell wall fraction of strain Guy11 might contribute to its efficient antifungal activity.
GluM attacks cell walls of M. oryzae and reduces virulence
To evaluate the action of GluM on M. oryzae, hydrolysis of the β-1,6-glucanase GluM toward the strain Guy11 cell wall was morphologically investigated. The tightly organized cell wall of strain Guy11 was converted into loose sacculi after GluM hydrolysis. Loosely packed fibers were observed around the GluM-treated cells by TEM (Fig. 3a, c). SEM analysis showed that the hyphae of Guy11 were transformed into a grid-like pattern, and strain Guy11 conidia were in a shrunken state after GluM treatment (Fig. 3b, d). β-1,6-glucan is essential for the rigid structure of fungal cell walls [36], where it has a cross-linking function. Therefore, the disorganization by GluM hydrolysis contributed to the loose state of the treated fungal cell walls. The reduced cell survival rates of strain Guy11 after GluM treatments (Fig. S8a), which is caused by the destruction of the cell wall integrity, indicate that GluM could be a key CWDE in the disintegration of fungal cell walls.
Structural changes of the cell walls of M. oryzae Guy11 after GluM treatments. Representative TEM (a, c) and SEM (b, d) images of the cell walls of strain Guy11 mycelia and conidia after GluM treatment (3 U/mL). Black arrows indicate the normal (solid) or loose (dashed) status of cell walls after GluM treatments in TEM analysis. White arrows indicate cell wall surfaces in flat status (solid), grid-like pattern or shrunken status (dashed) after GluM treatments in SEM analysis. Scale bar: 200 nm for TEM (a, c), 2 μm (b) and 5 μm (d) for SEM
Considering decomposition of M. oryzae cell walls, the interactions of GluM and its target of attack was investigated. Analysis showed that β-1,6-linked polysaccharides were present in the ASDN fraction extracted from M. oryzae Guy11 mycelia, which accounted for about 20% of the total cell wall polysaccharides of strain Guy11 (Table S8). Furthermore, the ASDN fraction was also extracted from GluM-treated mycelia and was found to account for less than 5% of the total cell wall polysaccharides, indicating the loss of β-1,6-linked polysaccharides. HPLC analysis of the ASDN fraction from strain Guy11 demonstrated that the molecular weight of ASDN polysaccharides was reduced after GluM hydrolysis (Figs. S8b, c), which was similar to the observations made with yeast glucans (Fig. 2b).
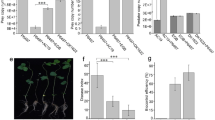
We further tested the effect of the β-1,6-glucanase on the germination of M. oryzae conidia and hyphal growth, which are important for the initial stage of rice seedling infection. Purified and expressed recombinant GluM exhibited inhibition of both germ tube and appressorium formation when compared to controls treated with TSUP (thermally inactivated SUP) and SUF (SUP ultrafiltrate) (Fig. 4a, b). The observed inhibition of conidial germination was dose-dependent, with a minimum effective GluM concentration of 1 U/mL (Fig. S9c). Higher doses of GluM decomposed conidial cell wall integrity completely (Fig. 4a). Time-dependent inhibition of mycelial growth was also observed (Fig. S9a). High turgor pressure of M. oryzae Guy11 appressoria is essential for the penetration of host cells. Therefore, the effect of GluM on the appressoria was investigated. In the presence of 2 M glycerol, the collapse rate of appressoria was about 57% following treatment with GluM, compared with 21% for the control experiment (Figs. 4c, Fig. S9b). Increasing the glycerol concentration to 4 M led to a disproportionate increase in collapsed appressoria to 89% after GluM treatment and to 47% for controls (Fig. 4c). This leads to fungal growth inhibition and inability to penetrate the host cell.
Effects of GluM on the germination of conidia, turgor pressure of appressoria and the virulence of strain Guy11. a Assays of conidial germination of wild-type Guy11, treatments including TSUP, SUF, SUP, GluM-S, and GluM-B (3 U/mL). The conidial germination (4 h) and appressoria formation (8 h) were observed using differential interference contrast (DIC) microscope, scale bars: 10 μm. b Quantification of the conidial germination and the appressorium formation. c Defective turgor pressure of appressoria in GluM-treated strain Guy11. The quantification of collapsed appressoria was performed by counting 100 appressoria. d Statistical analysis of the infectious growth of M. oryzae in rice sheath cells at 48 hpi after treatment with GluM at a dosage of 3, 6, and 12 U/mL. For each sample, appressorium penetration sites (n = 100) were observed and the invasive hyphae were classified according to type 1–4. Error bars denote st. dev. (SD), and values with different letters indicate statistically significant differences (p < 0.01). Scale bar: 10 μm. SUP: cell-free culture supernatants of strain EGB; TSUP: thermally inactivated SUP; SUF: SUP ultrafiltrate (<10 kDa); GluM-B: recombinant GluM from E. coli BL21 (DE3); GluM-S: purified GluM from SUP
To evaluate host invasion, we tested the effect of the β-1,6-glucanase on the penetration and invasive hyphal growth of strain Guy11 in rice sheath cells. The wild-type strain showed a 90% success rate of appressorium penetration events, with more than 80% classified as type 3 (extended but limited to one cell) and type 4 (extended to surrounding cells) invasive hyphae at 48 h post-inoculation (hpi) (Fig. 4d, Figs. S9d, e). In contrast, invasive hyphae were found at less than 40% of penetration sites following GluM treatment for all tested doses. Only 13% of appressoria had successfully penetrated the rice sheath cells after GluM treatment, and less than 5% of penetration sites showed type-3 and type-4 invasive growth (12 U/mL) (Fig. 4d). Obvious structural deformation was observed at 48 hpi, whereas invasive hyphae extended to the surrounding cells in the control (Fig. S9e). Furthermore, the overall infection severity of rice sheath cells by strain Guy11 could be reduced significantly in a dose-dependent manner by GluM. GluM also exhibited inhibitory activity against Fusarium graminearum, Ustilaginoidea virens and Botrytis cinerea (Fig. S10), indicating its potential broad-spectrum antifungal activity toward airborne plant-pathogenic fungi.
β-1,6-glucan is a main target in the control of fungi by strain EGB
Based on the fungal cell wall degrading ability of the β-1,6-glucanase GluM, we concluded that β-1,6-glucan structure is the main target in fungal predation by strain EGB. The relationship between predation by strain EGB and β-1,6-glucans in fungal cell walls was further investigated by exposing fungi with or without β-1,6-glucans in their cell wall to GluM. Both M. restricta and N. crassa were selected for predation experiments because their cell walls consist of 70% of β-1,6-glucan and free of β-1,6-glucans, respectively [23, 37]. Growth of M. restricta requires a relatively high proportion of olive oil in Dixon’s medium, which restricted the growth of strain EGB. Therefore, strain EGB was inoculated onto the colony when M. restricta was used as prey. In the case of N. crassa, strain EGB was not able to feed on the co-cultured N. crassa nor affect its growth and cell structure (Fig. 5a, Fig. S11a). At the observation time, abundant conidia formed and mycelia were rarely found. In contrast, the cellular morphology of M. restricta was changed at the contact sites with strain EGB. M. restricta cells were decomposed and strain EGB cells grew abundantly at inoculation sites (Fig. 5b, Figs. S11b, 12). Survival rates of N. crassa spores and M. restricta cells were measured. No obvious effects on the viability were observed for N. crassa, whereas M. restricta cells were decomposed and cell viability reduced to 21% after 10 h of GluM treatment (Fig. 5c, d). These results suggest that GluM specifically attacks β-1,6-glucans of fungal cell walls during predation.
Relationship between cell wall β-1,6-glucan contents and predatory behavior of strain EGB. a, b SEM analysis of the cell morphology. Strain EGB was co-cultured with N. crassa or M. restricta, and the cell morphologies of the fungi were observed. Representative SEM images are shown; scale bars: 2 μm. The dashed arrow indicates the contact site of strain EGB with M. restricta or N. crassa. c DIC images of the fungal cells. Representative DIC micrographs are shown; scale bars: 10 μm. d Survival rates of N. crassa spores and M. restricta cells after GluM treatments (3 U/mL). The quantitative data represent the means of three biological replicates. Error bars denote st. dev. (SD), and values with different letters indicate statistically significant differences (p < 0.01)
GluM is an essential weapon for the efficient predation of fungi by myxobacteria
From the results described above, the identified β-1,6-glucanase GluM from Corallococcus sp. strain EGB exhibited efficient fungal cell wall decomposing ability of β-1,6-glucan-containing fungal strains. Bioinformatics analysis revealed that GluM homologs are widely present in the sequenced genomes of myxobacteria, including the genera Myxococcus, Hyalangium, Stigmatella, Archangium, Melittangium, Cystobacter, Chondromyces and Sorangium (Table S7). We therefore deduced that this type of β-1,6-glucanase is an important weapon for efficient predation of fungi by myxobacteria. Because all attempts to genetically manipulate strain EGB failed, the model strain M. xanthus DK1622 bearing a GluM homolog Oar was tested as a substitute for strain EGB to determine the role of GluM in the predation of fungi by myxobacteria. After confirming that recombinant Oar also posesses β-1,6-glucanase activity (Tables S6 and 9), the Δoar mutant strain CL1001 and gluM complementation strain CL1002 (Δoar/gluM) were constructed. Due to the antifungal myxalamids secreted by strain DK1622 [38], the colonies of strain Guy11 were confined by strains DK1622, CL1001 and CL1002 (Fig. S13a). However, fractured mycelia were observed only with strains DK1622 and CL1002, which was in contrast to normal mycelium morphology observed with the control and strain CL1001 (Fig. S13b). Our SEM observations revealed that mycelial cell wall integrity of strain Guy11 treated with strain CL1001 was intact, compared with the perforated structure after treatments with strains DK1622 and CL1002, where the myxobacterial cells attached (Fig. 6).
Essential roles of GluM and Oar in the decomposition of strain Guy11 mycelia. Strains DK1622, CL1001, and CL1002 were co-cultured with strain Guy11. SEM analysis showing perforated structures at the contact sites of the cells of strains DK1622 and CL1002 (dashed arrow). Mycelial ell wall integrity was intact when co-cultured with strain CL1001 (solid arrow). Representative SEM images are shown; scale bars: 5 μm
To examine the function of the β-1,6-glucanase GluM in prey-predator interaction, strains DK1622, CL1001, CL1002 and EGB were individually co-cultured with M. oryzae Guy11 on TPM plates by spotting the myxobacterial cells next to the strain Guy11 colony. The interactions between strain Guy11 and myxobacteria were examined by time-lapse video microscopy (Movies S1–4). Otherwise, the growths of strains DK1622 and EGB were also observed with absence of strain Guy11 (Movies S5, 6). All myxobacterial strains moved in small cell aggregation with continuous fusion and separation as observed by time-lapse microscopy. Both strains DK1622 and EGB were able to penetrate into the colony of strain Guy11 within about 3 days, whereas strain EGB was able to move faster within the fungal colony (Figs. S14 a, b). Once approaching the mycelia of M. oryzae Guy11, strains DK1622 (Movie S1) and EGB (Movie S2) attached to and moved along the mycelia with swarming motility of 0.18 and 0.26 cm/day (Figs. S14a, b), respectively. The motility assay showed that motility of M. xanthus was not affected by Oar (data not shown). However, Δoar strain CL1001 lost the ability to perceive and move along the mycelia, even though the mycelia were in contact with the cell aggregations (Figs. S14a, b, Movie S3). Moreover, GluM complementation recovered the ability of strain CL1001 (Δoar) to perceive, move along and prey on M. oryzae Guy11 (Figs. S14a, b, Movie S4). Because TPM starvation agar is commonly used for developmental analysis of myxobacteria, mxyobacterial cell numbers decreased during incubation on TPM plates. However, upon contact with fungal mycelia, growth was promoted. Our qPCR analysis showed that the relative biomass of strains DK1622 and CL1002 increased by 90 and 130% from the predation of strain Guy11, respectively (Fig. S14c). However, the relative biomass of strain CL1001 (Δoar) decreased by 92% considering its developmental defect in myxospores formation [39] and cell lysis during incubation (Fig. S14c).
Discussion
Myxobacteria are ubiquitous micropredators that prey on a variety of other microorganisms by using a coordinated feeding strategy in microbial communities [40, 41]. Various species of Myxococcus, Sorangium, and Corallococcus have been shown to prey on several fungi and bacteria [42], and Corallococcus sp. strain EGB preyed more efficiently on fungi because it could grow and multiply at the cost of fungal biomass faster than strain DK1622 or other soil myxobacterial isolates (Fig. 1b, Figs. S14c, 15). During the predation event, antibiotics and bacteriolytic and proteolytic enzymes have been implicated in lysing prey cells [5]. Here, we show that a novel outer membrane β-1,6-glucanase contributes to predation of fungi by myxobacteria. This β-1,6-glucanase also mediates the sensing of fungi during predation and is therefore causal reason for killing prey.
β-barrel proteins are mainly found in the OM of Gram-negative bacteria [43], where they play crucial roles in signal transduction, metabolite transport and protein translocation, and account for 2–3% of proteins encoded in Gram-negative bacterial genomes [44]. GluM shared no identity with any known GH families, but shared high identity with the TonB-dependent receptor (Oar) of M. xanthus DK1622 [39]. Western blot analysis revealed that GluM is located in the bacterial membrane and computational structure analysis predicted multiple transmembrane domains and an overall barrel shape formed by β-sheets (Fig. S4). GluM was purified from the culture supernatant, and we deduced that it was released from the OM as part of OMVs, in which it has been detected by proteomic analysis [15]. Functional proteins that are associated with the OM are usually known as autotransporters, with only a few examples described to have protease or lipase activity. For example, the protease OmpT and phospholipase OMPLA from E. coli, as well as the deacylase LpxR from Salmonella typhimurium and LpxQ from Rhizobium leguminosarum [45, 46], were described to be important for various cellular processes including host immune defenses and biofilm formation [17]. The GluM homolog Oar was found to be involved in the development of M. xanthus DK1622 via transportation of the protease PopC [39, 47]. Here we found that GluM is also involved in the interaction of myxobacteria with fungi. As the first reported OM glycosidase, the identification of GluM expands the functional diversity of OM proteins to highlight another mechanism in predator-prey interactions.
The development of fungal antagonistic strategies for bacteria has created opportunities to establish new niches in terrestrial ecosystems [48]. Both Burkholderia and Serratia have evolved type III and type IV systems to deliver toxic effectors into competing fungal cells [49, 50]. The wide distribution of the secreted OM β-1,6-glucanase in myxobacteria represents another cross-kingdom interaction strategy between bacteria and fungi. Myxobacteria are ubiquitous in the environment, and we propose that β-1,6-glucanases in outer membrane vesicles are beneficial for engaging with fungi in a hostile relationship within soil environments. This hypothesis is supported by the observation that, in other genera of myxobacteria, homologs of GluM are widely distributed (Table S7), which is in contrast to the rare occurrence of β-1,3-glucanases in myxobacteria (Table S10). No β-1,3-glucanase activity was detected from the supernatant of strain EGB, and only negligible enzymatic and antifungal β-1,3-glucanase activities were previously reported in strain EGB [34]. We predict that the impact of fungi on myxobacterial evolution led to occupation of a specific ecological niche by myxobacteria using a OM β-1,6-glucanase as a targeted weapon against fungi.
Fungal diseases are a serious threat to overall plant agriculture, and therefore various antifungal agents have been researched. Considering the indispensable functions of the fungal cell wall in the survival, growth, development and pathogenicity, this aspect of fungal cells represents a highly attractive target for anti-fungal agent development [6]. The primary components of the fungal cell wall are β-glucans (β-1,3-glucan and β-1,6-glucan), α-glucans (α-1,3-glucan and α-1,4-glucan), chitin and mannan [51]. Corresponding β-1,3-glucanases, α-1,3-glucanase and chitinases have been investigated for their biocontrol potential against plant pathogens [9]. However, mutations or inhibition of the β-1,3-glucan biosynthesis can be complemented by an increase of the chitin content and the cross-linking of chitin to the β-1,6-glucan to maintain the insolubility and integrity of the wall [36]. Because β-1,6-glucan is only found in the cell walls of fungi and some members of oomycetes [52], it is a highly selective target for fungal killing. The roles of β-1,6-glucan have been investigated in S. cerevisiae, where it acts as a flexible glue by forming covalent cross-links between β-1,3-glucan, chitin and cell wall mannoproteins to create a rigid network [52, 53]. The cell wall of mutants defective in genes for β-1,6-glucan biosynthesis (kre6D, Kre6 and Skn1) shared the same features to cell walls defective in GluM-treated S. cerevisiae cells (Fig. S16)[11, 12, 54]. These data indicate that the β-(1,6)-glucan might be a more sensitive target for the control of pathogenic fungi compared to chitin or β-(1,3)-glucan. However, the ecological impacts of GluM on the non-pathogenic commensal fungal soil flora, which may also contain β-1,6-glucan, must be considered for further research.
The most efficient CWDE described so far is an acidic β-1,3-glucanase purified from potato, which inhibited the germination of Phytophthora infestans sporangia at a concentration of 0.3 μg/mL [55]. In contrast, GluM was capable of efficiently decomposing the mycelium and conidia of M. oryzae at a minimum concentration of 0.03 µg/mL (0.6 U/mL) (Fig. S9c). Although lyticase can completely degrade the cell wall of yeast, one of its major components, β-1,3-glucanase, cannot hydrolyze the β-1,3-glucan within the cell wall alone [56]. Chitinase E from Dioscorea opposita was able to control powdery mildew on strawberries at 0.1 mg/mL, and zymolyase showed no effect at a concentration of 6 mg/mL [57], whereas GluM effectively controlled rice blast disease at nM concentrations in our study (Fig. 4, Fig. S9). Direct observations of the hydrolyzed cell walls showed that those treated with GluM changed significantly compared to controls treated with other CWDEs. Only local degradation or shrinkage of the mycelium could be observed after treatment with chitinase or β-1,3-glucanase [58, 59].
Several functional β-1,6-glucanases, which are structurally unrelated to GluM, were identified in fungi. Their physiological roles were also attributed to parasitism and nutritional supply [13]. A comparison of the antifungal activity of GluM with that of β-1,6-glucanase from Trichoderma virens showed that GluM exhibited efficient decomposition at low dosages, which was not observed for T. virens β-1,6-glucanase even at high concentrations (Fig. S17). The highly effective fungal cell wall decomposition ability of GluM revealed in this work indicates a distinct role of myxobacterial β-1,6-glucanase when compared to previously reported β-1,6-glucanases, β-1,3-glucanases and chitinases, and enables strain EGB to effectively prey on a fungal pathogens. Overall, the identification and functional characterization of OM β-1,6-glucanase provide new implications for understanding the molecular ecological basis of myxobacteria-fungi interaction.
References
Munozdorado J, Marcostorres FJ, Garciabravo E, Moraledamunoz A, Perez J. Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol. 2016;7:781–781.
Keane R, Berleman JE. The predatory life cycle of Myxococcus xanthus. Microbiology. 2016;162:1–11.
Livingstone PG, Millard AD, Swain MT, Whitworth DE. Transcriptional changes when Myxococcus xanthus preys on Escherichia coli suggest myxobacterial predators are constitutively toxic but regulate their feeding. Microb Genom. 2018;4:e000152.
Evans AG, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, et al. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology. 2012;158:2742–52.
Morgan AD, MacLean RC, Hillesland KL, Velicer GJ. Comparative analysis of Myxococcus predation on soil bacteria. Appl Environ Microbiol. 2010;76:6920–7.
Latgé JP. The cell wall: a carbohydrate armour for the fungal cell. Mol Microbiol. 2007;66:279–90.
Mauch F, Mauchmani B, Boller T. Antifungal hydrolases in pea tissue: II. Inhibition of fungal growth by combinations of chitinase and β-1,3-glucanase. Plant Physiol. 1988;88:936–42.
Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species-opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56.
Selitrennikoff CP. Antifungal proteins. Appl Environ Microbiol. 2001;67:2883–94.
Umeyama T, Kaneko A, Watanabe H, Hirai A, Uehara Y, Niimi M, et al. Deletion of the CaBIG1 gene reduces β-1, 6-glucan synthesis, filamentation, adhesion, and virulence in Candida albicans. Infect Immun. 2006;74:2373–81.
Gilbert NM, Donlin MJ, Gerik KJ, Specht CA, Djordjevic JT, Wilson CF, et al. KRE genes are required for β-1, 6-glucan synthesis, maintenance of capsule architecture and cell wall protein anchoring in Cryptococcus neoformans. Mol Microbiol. 2010;76:517–34.
Oliveira-GarciaE, Deising HB. Attenuation of PAMP-triggered immunity in maize requires down-regulation of the key β-1, 6-glucan synthesis genes KRE5 and KRE6 in biotrophic hyphae of Colletotrichum graminicola. Plant J. 2016;87:355–75.
Djonović S, Pozo MJ, Kenerley CM. Tvbgn3, a β-1, 6-glucanase from the biocontrol fungus Trichoderma virens, is involved in mycoparasitism and control of Pythium ultimum. Appl Environ Microbiol. 2006;72:7661–70.
Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol. 2015;13:605–19.
Berleman JE, Allen S, Danielewicz MA, Remis JP, Gorur A, Cunha J, et al. The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol. 2014;5:474.
Chaturvedi D, Mahalakshmi R. Transmembrane β-barrels: evolution, folding and energetics. BBA-Biomembranes. 2017;1859:2467–82.
Bishop RE. Structural biology of membrane-intrinsic β-barrel enzymes: Sentinels of the bacterial outer membrane. BBA-Biomembranes. 2008;1778:1881–96.
Pimentel D, McNair S, Janecka J, Wightman J, Simmonds C, O’connell C, et al. Economic and environmental threats of alien plant, animal, and microbe invasions. Agr Ecosyst Environ. 2001;84:1–20.
Glare T, Caradus J, Gelernter W, Jackson T, Keyhani N, Köhl J, et al. Have biopesticides come of age? Trends Biotechnol. 2012;30:250–8.
Bull CT, Shetty KG, Subbarao KV. Interactions between myxobacteria, plant pathogenic fungi, and biocontrol agents. Plant Dis. 2002;86:889–96.
Li Z, Ye X, Chen P, Ji K, Zhou J, Wang F, et al. Antifungal potential of Corallococcus sp. strain EGB against plant pathogenic fungi. Biol Control. 2017;110:10–17.
Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74:2938–42.
Stalhberger T, Simenel C, Clavaud C, Eijsink VG, Jourdain R, Delepierre M, et al. Chemical organization of the cell wall polysaccharide core of Malassezia restricta. J Biol Chem. 2014;289:12647–56.
Zhong K, Li X, Le X, Kong X, Zhang H, Zheng X, et al. MoDnm1 dynamin mediating peroxisomal and mitochondrial fission in complex with MoFis1 and MoMdv1 is important for development of functional appressorium in Magnaporthe oryzae. PLoS Pathog. 2016;12:e1005823.
Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–90.
Qi Z, Liu M, Dong Y, Zhu Q, Li L, Li B, et al. The syntaxin protein (MoSyn8) mediates intracellular trafficking to regulate conidiogenesis and pathogenicity of rice blast fungus. New Phytol. 2016;209:1655–67.
Valent B, Farrall L, Chumley FG. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics. 1991;127:87–101.
Park DS, Sayler RJ, Hong YG, Nam MH, Yang Y. A method for inoculation and evaluation of rice sheath blight disease. Plant Dis. 2008;92:25–29.
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem. 1959;31:426–426.
Li Z, Wu J, Zhang B, Wang F, Ye X, Huang Y, et al. AmyM, a novel maltohexaose-forming α-amylase from Corallococcus sp. strain EGB. Appl Environ Microbiol. 2015;81:1977–87.
Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67.
Bi H, Ni X, Liu X, Iteku J, Tai G, Zhou Y, et al. A novel water-soluble β-(1 → 6)-D-glucan isolated from the fruit bodies of Bulgaria inquinans (Fries). Carbohyd Res. 2009;344:1254–8.
Xiu A, Kong Y, Zhou M, Zhu B, Wang S, Zhang J. The chemical and digestive properties of a soluble glucan from Agrobacterium sp. ZX09. Carbohyd Polym. 2010;82:623–8.
Zhou J, Li Z, Wu J, Li L, Li D, Ye X, et al. Functional analysis of a novel β-(1,3)-glucanase from Corallococcus sp. EGB containing a fascin-like module. Appl Environ Microbiol. 2017;83:e01016–17.
Ebbole DJ. Magnaporthe as a model for understanding host-pathogen interactions. Annu Rev Phytopathol. 2007;45:437–56.
Lipke PN, Ovalle R. Cell wall architecture in yeast: new structure and new challenges. J Bacteriol. 1998;180:3735–40.
Maddi A, Dettman A, Fu C, Seiler S, Free SJ. WSC-1 and HAM-7 are MAK-1 MAP kinase pathway sensors required for cell wall integrity and hyphal fusion in Neurospora crassa. PLoS ONE. 2012;7:e42374.
Gerth K, Jansen R, Reifenstahl G, Höfle G, Irschik H, Kunze B, et al. The myxalamids, new antibiotics from Myxococcus xanthus (Myxobacterales). J Antibiot. 1983;36:1150–6.
Martinez-Canamero M, Munoz-Dorado J, Farez-Vidal E, Inouye M, Inouye S. Oar, a 115-kilodalton membrane protein required for development of Myxococcus xanthus. J Bacteriol. 1993;175:4756–63.
Hillesland KL, Lenski RE, Velicer GJ. Ecological variables affecting predatory success in Myxococcus xanthus. Microb Ecol. 2007;53:571–8.
Zhou XW, Li SG, Li W, Jiang DM, Han K, Wu ZH, et al. Myxobacterial community is a predominant and highly diverse bacterial group in soil niches. Environ Microbiol Rep. 2014;6:45–56.
Dahm M, Brzezinska AJ, Wrótniak-Drzewiecka W, Golinska P, Rozycki H, Rai M. Myxobacteria as a potential biocontrol agent effective against pathogenic fungi of economically important forest trees. Dendrobiology. 2015;74:13–24.
Hagan CL, Silhavy TJ, Kahne D. β-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210.
Wenz L-S, Qiu J, Becker T, Wiedemann N. Biogenesis and folding of β-barrel membrane proteins. Cell Cycle. 2014;13:169–70.
Reynolds CM, Ribeiro AA, McGrath SC, Cotter RJ, Raetz CR, Trent MS. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J Biol Chem. 2006;281:21974–87.
Que-Gewirth NL, Karbarz MJ, Kalb SR, Cotter RJ, Raetz CR. Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase lpxQ. J Biol Chem. 2003;278:12120–9.
Gómez-Santos N, Glatter T, Koebnik R, Świątek-Połatyńsk M A, Søgaard-Andersen L. A TonB-dependent transporter is required for secretion of protease PopC across the bacterial outer membrane. Nat Commun. 2019;10:1360-1377.
Wd Boer, Folman LB, Summerbell RC, et al. in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811.
Swain DM, Yadav SK, Tyagi I, Kumar R, Kumar R, Ghosh S, et al. A prophage tail-like protein is deployed by Burkholderia bacteria to feed on fungi. Nat Commun. 2017;8:404.
Trunk K, Peltier J, Liu YC, Dill BD, Walker L, Gow N, et al. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 2018;3:920–31.
Synytsya A, Novák M. Structural diversity of fungal glucans. Carbohyd Polym. 2013;92:792–809.
Fesel PH, Zuccaro A. β-glucan: crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet Biol. 2016;90:53–60.
Klis FM, Boorsma A, De Groot PW. Cell wall construction in Saccharomyces cerevisiae. Yeast. 2006;23:185–202.
Osumi M. The ultrastructure of yeast: cell wall structure and formation. Micron. 1998;29:207–33.
Tonon C, Guevara G, Oliva C, Daleo G. Isolation of a potato pcidic 39 kDa β-1, 3-glucanase with antifungal activity against Phytophthora infestans and analysis of its expression in potato cultivars differing in their degrees of field resistance. J Phytopathol. 2002;150:189–95.
Scott JH, Schekman R. Lyticase: endoglucanase and protease activities that act together in yeast cell lysis. J Bacteriol. 1980;142:414–23.
Karasuda S, Tanaka S, Kajihara H, Yamamoto Y, Koga D. Plant chitinase as a possible biocontrol agent for use instead of chemical fungicides. Biosci Biotechnol Biochem. 2003;67:221–4.
Fridlender M, Inbar J, Chet I. Biological control of soilborne plant pathogens by a β-1, 3 glucanase-producing Pseudomonas cepacia. Soil Biol Biochem. 1993;25:1211–21.
Benhamou N, Broglie K, Broglie R, Chet I. Antifungal effect of bean endochitinase on Rhizoctonia solani: ultrastructural changes and cytochemical aspects of chitin breakdown. Can J Microbiol. 1993;39:318–28.
Acknowledgements
We would like to thank Prof. Xiaohua Mao and Prof. Huiqiang Lou for generously providing strains Myxococcus xanthus DK1622 and Neurospora crassa, Prof. Yifa Zhou for generously providing the β-1,6-glucan BIWP2, Prof. Yuezhong Li for the kind suggestions for gene knockout, Prof. Yufeng Yao and Prof. Daniel Wall and Prof. Ruifu Zhang for helpful manuscript suggestions. We are grateful to Prof. Josef Voglmeir for revising the manuscript. This work was supported by the Major State Basic Research Development Program of China (973 program, no. 2015CB150502), the National Natural Science Foundation of China (No. 31570059 and 31700054), the Postdoctoral Science Foundation of China (No. 2016M591859) and Jiangsu Agricultural science and technology innovation fund (CX(18)1003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Z., Ye, X., Liu, M. et al. A novel outer membrane β-1,6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J 13, 2223–2235 (2019). https://doi.org/10.1038/s41396-019-0424-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-019-0424-x
This article is cited by
-
Glucanase Gene of Trichoderma; New Strategy for the Management of Root Rot Disease in Chili
Journal of Soil Science and Plant Nutrition (2024)
-
Myxobacteria restrain Phytophthora invasion by scavenging thiamine in soybean rhizosphere via outer membrane vesicle-secreted thiaminase I
Nature Communications (2023)
-
Predation of oomycetes by myxobacteria via a specialized CAZyme system arising from adaptive evolution
The ISME Journal (2023)
-
A myxobacterial GH19 lysozyme with bacteriolytic activity on both Gram-positive and negative phytopathogens
AMB Express (2022)
-
Conserved and reproducible bacterial communities associate with extraradical hyphae of arbuscular mycorrhizal fungi
The ISME Journal (2021)