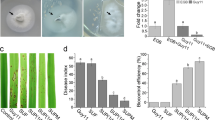
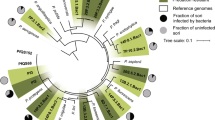
Abstract
As social micropredators, myxobacteria are studied for their abilities to prey on bacteria and fungi. However, their predation of oomycetes has received little attention. Here, we show that Archangium sp. AC19 secretes a carbohydrate-active enzyme (CAZyme) cocktail during predation on oomycetes Phytophthora. These enzymes include three specialized β-1,3-glucanases (AcGlu13.1, –13.2 and –13.3) that act as a cooperative consortium to target β-1,3-glucans of Phytophthora. However, the CAZymes showed no hydrolytic effects on fungal cells, even though fungi contain β-1,3-glucans. Heterologous expression of AcGlu13.1, –13.2 or –13.3 enzymes in Myxococcus xanthus DK1622, a model myxobacterium that antagonizes but does not predate on P. sojae, conferred a cooperative and mycophagous ability that stably maintains myxobacteria populations as a mixture of engineered strains. Comparative genomic analyses suggest that these CAZymes arose from adaptive evolution among Cystobacteriaceae myxobacteria for a specific prey killing behavior, whereby the presence of Phytophthora promotes growth of myxobacterial taxa by nutrient release and consumption. Our findings demonstrate that this lethal combination of CAZymes transforms a non-predatory myxobacterium into a predator with the ability to feed on Phytophthora, and provides new insights for understanding predator-prey interactions. In summary, our work extends the repertoire of myxobacteria predatory strategies and their evolution, and suggests that these CAZymes can be engineered as a functional consortium into strains for biocontrol of Phytophothora diseases and hence crop protection.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
We are sorry, but there is no personal subscription option available for your country.
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hungate BA, Marks JC, Power ME, Schwartz E, van Groenigen KJ, Blazewicz SJ, et al. The functional significance of bacterial predators. mBio. 2021;12:e00466–00421.
Zhou X, Li S, Li W, Jiang D, Han K, Wu Z, et al. Myxobacterial community is a predominant and highly diverse bacterial group in soil niches. Env Microbiol Rep. 2014;6:45–56.
Ye X, Li Z, Luo X, Wang W, Li Y, Li R, et al. A predatory myxobacterium controls cucumber Fusarium wilt by regulating the soil microbial community. Microbiome. 2020;8:49.
Petters S, Groß V, Söllinger A, Pichler M, Reinhard A, Bengtsson M, et al. The soil microbial food web revisited: Predatory myxobacteria as keystone taxa? ISME J. 2021;15:2665–75.
Zhang W, Wang Y, Lu H, Liu Q, Wang C, Hu W, et al. Dynamics of solitary predation by Myxococcus xanthus on Escherichia coli observed at the single-cell level. Appl Environ Microbiol. 2020;86:e02286–02219.
Muñoz-Dorado J, Marcos-Torres FJ, García-Bravo E, Moraleda-Muñoz A, Pérez J. Myxobacteria: moving, killing, feeding, and surviving together. Front Microbiol. 2016;7:781.
Li Z, Ye X, Liu M, Xia C, Zhang L, Luo X, et al. A novel outer membrane β-1, 6-glucanase is deployed in the predation of fungi by myxobacteria. ISME J. 2019;13:2223–35.
Kamoun S. Molecular genetics of pathogenic oomycetes. Eukaryot Cell. 2003;2:191–9.
Berleman JE, Kirby JR. Deciphering the hunting strategy of a bacterial wolfpack. FEMS Microbiol Rev. 2009;33:942–57.
Xiao Y, Wei X, Ebright R, Wall D. Antibiotic production by myxobacteria plays a role in predation. J Bacteriol. 2011;193:4626–33.
Seef S, Herrou J, de Boissier P, My L, Brasseur G, Robert D, et al. A Tad-like apparatus is required for contact-dependent prey killing in predatory social bacteria. Elife 2021;10:e72409.
Thiery S, Turowski P, Berleman JE, Kaimer C. The predatory soil bacterium Myxococcus xanthus combines a Tad-and an atypical type 3-like protein secretion system to kill bacterial cells. Cell Rep. 2022;40:111340.
Grenville-Briggs LJ, Anderson VL, Fugelstad J, Avrova AO, Bouzenzana J, Williams A, et al. Cellulose synthesis in Phytophthora infestans is required for normal appressorium formation and successful infection of potato. Plant Cell. 2008;20:720–38.
Fesel PH, Zuccaro A. β-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet Biol. 2016;90:53–60.
Tonón C, Daleo G, Oliva C. An acidic β-1, 3 glucanase from potato tubers appears to be patatin. Plant Physiol Bioch. 2001;39:849–54.
Wang Y, Tyler BM, Wang Y. Defense and counterdefense during plant-pathogenic oomycete infection. Annu Rev Microbiol. 2019;73:667–96.
Clavaud C, Aimanianda V, Latge J-P Organization of fungal, oomycete and lichen (1, 3)-β-glucans. Chemistry, Biochemistry, and Biology of 1-3 Beta Glucans and Related Polysaccharides: Elsevier; 2009. p. 387-424.
Liu N, Li H, Chevrette MG, Zhang L, Cao L, Zhou H, et al. Functional metagenomics reveals abundant polysaccharide-degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite. ISME J. 2019;13:104–17.
Feng H, Wan C, Zhang Z, Chen H, Li Z, Jiang H, et al. Specific interaction of an RNA-binding protein with the 3′-UTR of its target mRNA is critical to oomycete sexual reproduction. PLoS Pathog. 2021;17:e1010001.
Li Z, Ye X, Chen P, Ji K, Zhou J, Wang F, et al. Antifungal potential of Corallococcus sp. strain EGB against plant pathogenic fungi. Biol Control. 2017;110:10–7.
Li Z, Wang T, Luo X, Li X, Xia C, Zhao Y, et al. Biocontrol potential of Myxococcus sp. strain BS against bacterial soft rot of calla lily caused by Pectobacterium carotovorum. Biol Control 2018;126:36–44.
Wardman JF, Bains RK, Rahfeld P, Withers SG. Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat Rev Microbiol. 2022;20:542–56.
Rakoff-Nahoum S, Coyne M, Comstock L. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24:40–9.
Gong Y, Zhang Z, Liu Y, Zhou XW, Anwar MN, Li Z, et al. A nuclease‐toxin and immunity system for kin discrimination in Myxococcus xanthus. Environ Microbiol. 2018;20:2552–67.
Gao C, Xu H, Huang J, Sun B, Zhang F, Savage Z, et al. Pathogen manipulation of chloroplast function triggers a light-dependent immune recognition. Proc Natl Acad Sci USA. 2020;117:9613–20.
Huang J, Gu L, Zhang Y, Yan T, Kong G, Kong L, et al. An oomycete plant pathogen reprograms host pre-mRNA splicing to subvert immunity. Nat Commun. 2017;8:1–15.
Xiao K, Kinkel LL, Samac DA. Biological control of Phytophthora root rots on alfalfa and soybean with Streptomyces. Biol Control. 2002;23:285–95.
Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Biochem. 1976;72:248–54.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Wawra S, Fesel P, Widmer H, Neumann U, Lahrmann U, Becker S, et al. FGB1 and WSC3 are in planta‐induced β‐glucan‐binding fungal lectins with different functions. N. Phytol. 2019;222:1493–506.
Ma Z, Song T, Zhu L, Ye W, Wang Y, Shao Y, et al. A Phytophthora sojae glycoside hydrolase 12 protein is a major virulence factor during soybean infection and is recognized as a PAMP. Plant Cell. 2015;27:2057–72.
Furness E, Whitworth DE, Zwarycz A Predatory interactions between myxobacteria and their prey. The Ecology of Predation at The Microscale: Springer. p. 2020;1-36.
Tyler BM. Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol Plant Pathol. 2007;8:1–8.
Zhou J, Li Z, Wu J, Li L, Li D, Ye X, et al. Functional analysis of a novel β-(1, 3)-glucanase from Corallococcus sp. strain EGB containing a fascin-like module. Appl Environ Microbiol. 2017;83:e01016–17.
Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Curr Opin Genet Dev. 2005;15:589–94.
Pérez J, Moraleda‐Muñoz A, Marcos‐Torres FJ, Muñoz‐Dorado J. Bacterial predation: 75 years and counting! Environ Microbiol. 2016;18:766–79.
Evans R, Beckerman AP, Wright RC, McQueen-Mason S, Bruce NC, Brockhurst MA. Eco-evolutionary dynamics set the tempo and trajectory of metabolic evolution in multispecies communities. Curr Biol. 2020;30:4984–8.e4
Bretscher AP, Kaiser D. Nutrition of Myxococcus xanthus, a fruiting myxobacterium. J Bacteriol. 1978;133:763–8.
Wang J, Wang J, Wu S, Zhang Z, Li Y. Global geographic diversity and distribution of the Myxobacteria. Microbiol Spectr. 2021;9:e00012–21.
Waite DW, Chuvochina M, Pelikan C, Parks DH, Yilmaz P, Wagner M, et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int J Syst Evol Microbiol. 2020;70:5972–6016.
Thiery S, Kaimer C. The predation strategy of Myxococcus xanthus. Front Microbiol. 2020;11:2.
Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 2010;330:831–5.
Irschik H, Reichenbach H. An unusual pattern of carbohydrate utilization in Corallococcus (Myxococcus) coralloides (Myxobacterales). Arch Microbiol. 1985;142:40–4.
Cao P, Dey A, Vassallo CN, Wall D. How myxobacteria cooperate. J Mol Biol. 2015;427:3709–21.
Rosenberg E, Keller KH, Dworkin M. Cell density-dependent growth of Myxococcus xanthus on casein. J Bacteriol. 1977;129:770–7.
Marshall RC, Whitworth DE. Is “Wolf‐Pack” predation by antimicrobial bacteria cooperative? Cell behaviour and predatory mechanisms indicate profound selfishness, even when working alongside kin. BioEssays. 2019;41:1800247.
Acknowledgements
We thank Professor Daolong Dou and Suomeng Dong (Nanjing Agricultural University) for generously providing Phytophthora strains, Assistant Professor Danyu Shen (Nanjing Agricultural University) and Junjun Wu (Jiangnan University) for genome analysis, and Assistant Professor Xu Han (Tianjin Institute of Industrial Biotechnology Chinese Academy of Sciences) for protein structure predictions. This work was supported by the National Natural Science Foundation of China (32070027 to ZL, 32170123 to ZC and 32270066 to ZL), the Key Research & Development Plan of Jiangsu Province (BE2020340 to ZC), the National Science and Technology Major Project (2020ZX08009-04B to ZC), the National Key Research and Development Program of China (2021YFC2103600 to ZC and ZL) and the Fundamental Research Funds for the Central Universities (KYZZ2022001 to ZL). We also thank Mr. Gang Hu and Chun Qin (Electron Microscope Experimental Center, College of Life Sciences, Nanjing Agricultural University) for their help with SEM.
Author information
Authors and Affiliations
Contributions
Z.L., L.Z., and Z.C. designed the experiments; L.Z. performed the majority of the experiments and data interpretation. JHW and CD expressed enzymes and performed pot experiments. ML, JYW, JH and XL performed binding and infection assays; LL, CX and LLZ. helped with experiments. XY and YH analyzed the data; JJW, YL and DW contributed in myxobacterial isolates, evolutionary analysis and manuscript edits. LZ, ZKL and ZC wrote and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Dong, C., Wang, J. et al. Predation of oomycetes by myxobacteria via a specialized CAZyme system arising from adaptive evolution. ISME J 17, 1089–1103 (2023). https://doi.org/10.1038/s41396-023-01423-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-023-01423-y