Abstract
The PHLDA3 gene encodes a small 127 amino acid protein with a pleckstrin homology (PH)-only domain. The expression and significance of PHLDA3 in lung cancer remain unclear. Here, we investigated the role of PHLDA3 in tumor proliferation and invasion in lung adenocarcinoma. Immunohistochemistry and immunoblotting analyses were used to assess PHLDA3 expression in lung cancer tissues, and its correlation with clinicopathological factors in lung cancer. Plasmids encoding PHLDA3 and small interfering RNA against PHLDA3 were used to regulate the expression of PHLDA3 in lung cancer cells. Furthermore, the effects of PHLDA3 on lung cancer cell proliferation and invasion were investigated using the MTS, colony formation, Matrigel invasion, and wound healing assays. Co-immunoprecipitation analysis and inhibitors of both the Wnt signaling pathway and GSK3β were used to explore the regulatory mechanisms underlying the role of PHLDA3 in lung cancer cells. PHLDA3 was found to be overexpressed in lung cancer tissues, and its expression was correlated with poor outcomes in lung adenocarcinoma patients. PHLDA3 expression promoted the proliferation, invasion, and migration of lung cancer cells. Overexpression of PHLDA3 activated the Wnt signaling pathway and facilitated epithelial–mesenchymal transition. Inhibition of Wnt signaling pathway activity, using XAV-939, reversed the effects of PHLDA3 overexpression in lung cancer cells; moreover, PHLDA3 could bind to GSK3β. Inhibition of GSK3β activity, using CHIR-99021, restored the proliferative and invasive abilities of PHLDA3 knockdown cells. Our findings demonstrate that PHLDA3 is highly expressed in lung adenocarcinomas and is correlated with poor outcomes. Furthermore, it promotes the proliferation and invasion of lung cancer cells by activating the Wnt signaling pathway.
Similar content being viewed by others
Introduction
The PHLDA3 gene, located on mouse chromosome 1 and human chromosome 1q31 [1], encodes a 127 amino acid protein with a pleckstrin homology (PH)-only domain [2]. The PH domain is an amino acid sequence of ~100 residues with a specific three-dimensional structure, allowing binding to phosphoinositides and protein–protein interactions. It is present in various proteins involved in signal transduction, phospholipid processing, membrane trafficking, and cytoskeletal organization [3]. PHLDA3 has been demonstrated to regulate acute kidney [4] and liver injury [5] induced by cisplatin and tunicamycin, respectively. Notably, the effects of PHLDA3 on islets and β cells are complex and varied. PHLDA3 deficiency improves islet engraftment by suppressing hypoxic damage [6], whereas PHLDA3 induction protects against cell death during stress [7]. Similar to this bidirectional regulation, PHLDA3 expression can ameliorate pressure overload-induced cardiac remodeling mainly by blocking the Akt signaling pathway [8]; additionally, its overexpression impairs the specification of hemangioblasts and vascular development in zebrafish [9]. As a direct target of p53, PHLDA3 acts as a tumor suppressor in neuroendocrine tumors by competitively binding to PIP to inhibit Akt activity [10,11,12]. In esophageal squamous cell carcinomas, low PHLDA3 expression is associated with poor prognosis [13], whereas PHLDA3 levels are significantly increased in head and neck squamous cell carcinoma [14]. Nevertheless, the role of PHLDA3 in lung cancer biology has been poorly studied and remains to be fully elucidated.
Lung cancer, of which 80–85% is non-small cell lung cancer (NSCLC), is the leading cause of cancer-related deaths worldwide [15]. Wnt/β-catenin alterations are prominent in human malignancies, and available data indicate that Wnt signaling substantially impacts NSCLC tumorigenesis, prognosis, and resistance to therapy [16]. This signaling pathway is regulated by a large and complex array of proteins, including stabilized cytoplasmic β-catenin regulated by the destruction complex, which plays a key role in signaling output of the canonical Wnt cascade. When the Fz/LRP receptors are not engaged, CK1 and GSK3 sequentially phosphorylate the axin-bound β-catenin at a series of regularly spaced N-terminal Ser/Thr residues, which results in its degradation via the ubiquitin-mediated proteasomal pathway. However, upon activation of the Wnt pathway, β-catenin dissociates from the destruction complex, accumulates in the cytoplasm, then enters the nucleus, associates with TCF4, and transforms the latter into a transcriptional activator that activates target genes, such as c-Myc (MYC), CyclinD1 (CCND1), and matrix metallopeptidase 7 (MMP7) in the Wnt pathway [17,18,19]. Epithelial–mesenchymal transition (EMT) is a fundamental biological process that occurs during normal organ development, tissue remodeling, and wound healing, and contributes pathologically to fibrosis and cancer progression. The canonical Wnt pathway has long been considered a key activator of EMT [20, 21].
In this study, we examined the expression of PHLDA3 in lung cancer tissues and corresponding normal lung tissues and analyzed its potential correlation with clinicopathological factors. We also explored the regulatory effects of PHLDA3 on the Wnt signaling pathway and EMT by increasing or decreasing the expression of PHLDA3 in lung cancer cells. Finally, we investigated the effects of PHLDA3 on the proliferative and invasive abilities of lung cancer cells.
Materials and methods
Cell culture and transfection
The human lung cancer cell lines A549 and H1299 were obtained from the Shanghai Cell Bank (Shanghai, China) and cultured according to the instructions of the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 (Gibco, Grand Island, NY, USA), and 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) was added at 37 °C in 5% CO2; the cells were grown in sterile culture dishes and passaged with 0.25% trypsin (Gibco, Carlsbad, CA, USA) every 1 or 2 days.
For transfection, Lipofectamine 3000 (Invitrogen) transfection reagent was used to transfect plasmids and small interfering RNAs (siRNAs) according to the manufacturer’s instructions. pCMV6-Myc-DDK (#PS100001) and pCMV6-Myc-DDK-PHLDA3 (#RC206751) were purchased from Origene (Rockville, MD, USA). siRNA against PHLDA3 (siPHLDA3) and control scrambled siRNA were purchased from RiboBio (Guangzhou, China). The oligonucleotides used were as follows: siPHLDA3 1#, 5′-TGGTCAAGTTCAAGAACCA-3′; siPHLDA3 2#, 5′-GCCACATCTACTTCACGCT-3′; siPHLDA3 3#, 5′-GGCCCAAGGAGCTCAGCTT-3′.
An inhibitor of Wnt/β-catenin signaling, XAV-939, and the GSK3α/β inhibitor CHIR-99021 were purchased from MedChemExpress (Monmouth Junction, NJ, USA). The concentrations of XAV-939 and CHIR-99021 were 20 and 30 nM, respectively. Both XAV-939 and CHIR-99021 were dissolved in dimethyl sulfoxide (DMSO) (Beijing Solarbio Science & Technology Co., Beijing, China) and added 24 h after transfection (for 24 h); the same volume of DMSO was added to the control cells.
Patients and specimens
We collected 206 NSCLC specimens from the patients (average age of 58 years) who underwent complete surgical resection at the First Affiliated Hospital of China Medical University between 2012 and 2015. Some of the lung cancer samples (108 cases) were accompanied by corresponding normal lung tissue samples (>5 cm distal to the primary tumor edge). Written informed consent was obtained from all the patients, and all procedures were approved by the Research Ethics Committee of China Medical University. All tumor specimens were obtained during surgical resection, and all patients were chemotherapy- and radiotherapy-naïve prior to resection. The histological diagnoses and differentiation grades of the tissue samples were classified according to the World Health Organization classification system as squamous cell carcinoma (LUSC) (n = 98) and adenocarcinoma (LUAD) (n = 108) [22]. The cancers were classified as well (20 cases), moderately (136 cases), and poorly differentiated (50 cases). According to the seventh edition of the International Union against Cancer TNM Staging System for Lung Cancer [23], patients were categorized as stage I (n = 32), II (n = 133), III (n = 33), or IV (n = 8). Lymph node metastases were observed in 105 patients. We also collected 60 pairs of fresh primary lung cancer and matched normal lung tissue specimens. Samples were frozen in liquid nitrogen immediately after removal and stored at −80 °C.
Immunohistochemistry and immunofluorescence
Immunohistochemical assays were performed as described previously [24]. Briefly, tissue sections were incubated with an anti-PHLDA3 rabbit polyclonal antibody (ab196757, 1:100; Abcam, Cambridge, MA, USA). The intensity of PHLDA3 staining was scored as follows: 0, no staining; 1 weak; 2, moderate; and 3, high. Percentage scores were categorized as follows: 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The scores for each tumor sample were multiplied to obtain a final score ranging from 0 to 12. Tumor samples with scores ≥6 were considered to have positive expression, while those with scores ≥0 and <6 were considered to be negative for the expression of PHLDA3.
For immunofluorescence staining, cells were fixed, permeabilized, and incubated with primary antibodies and fluorescein isothiocyanate- or tetramethyl rhodamine isothiocyanate-conjugated secondary antibodies. Nuclei were counterstained with 4,6-diamidino-2-phenylindole, and the cells were observed under a confocal microscope.
Immunoblotting analysis and immunoprecipitation
The assays were performed as described previously [25]. Information regarding the primary antibodies used for immunoblotting is provided in Supplementary Table 1. Protein bands were visualized using enhanced chemiluminescence (Pierce, Thermo Fisher Scientific, Waltham, MA, USA) and detected using a bio-imaging system (DNR Bio-Imaging System, Jerusalem, Israel). Relative protein levels were calculated after normalization to the levels of GAPDH, β-actin, or laminB, used as the loading control. The bands were quantified using densitometry and the ImageJ software.
For immunoprecipitation, the lysates were mixed and incubated overnight with protein G-agarose beads (Bimake, Houston, TX) and the corresponding primary antibodies. The results were analyzed using western blotting.
MTS and colony formation assays
For the MTS (Promega, Madison, USA) assay, 24 h after transfection, the cells were seeded in 96-well plates (3000 cells/well) and incubated in a medium containing 10% FBS, for appropriate times. The MTS reagent (20 µl/well) was added for 1 h at 37 °C, and the optical density was measured at 490 nm every 24 h for 5 days to generate a growth curve.
For colony formation assays, cells were seeded into 6 cm2 culture dishes (500 cells/dish) 24 h after transfection and then incubated for 10–15 days. The culture medium was changed every 4–5 days. The cells were then washed with phosphate-buffered saline, fixed with 4% paraformaldehyde for 20 min, and stained with Giemsa solution for 10 min. The number of colonies with more than 50 cells was counted. All experiments were performed in triplicate.
Matrigel invasion and wound healing assays
Assays were performed as previously described [26]. For the invasion assays, we used a 24-well transwell chamber with a pore size of 8 μm (Costar, Cambridge, MA, USA), with inserts coated with 100 μl Matrigel (1:7 dilution, BD Bioscience). Twenty-four hours after transfection, we added 6–12 × 105 cells in 100 μl medium supplemented with 2% FBS to the upper Matrigel chamber and incubated for 24 h. Medium supplemented with 20% FBS was added to the lower chamber as a chemoattractant. The number of invading cells was counted in ten randomly selected fields under a microscope at a high magnification (×200).
For wound healing assays, cells were grown to confluence in 6-well plates, and the cell monolayer was scratched using a pipette tip. Wound healing within the scratch line was observed at the indicated time points, and representative scrape lines for each cell line were photographed. Duplicate wells for each condition were examined in each experiment. Wound dimensions were measured optically using Photoshop software. All experiments were performed in triplicate.
Statistical analysis
All statistical analyses were performed using the SPSS software (version 17.0; Chicago, IL, USA) and the GraphPad Prism software (version 6.0; La Jolla, CA, USA). Immunohistochemistry results were analyzed using the chi-square test and Spearman’s rank correlation test. Differences between groups were compared using a two-tailed Student’s t test and considered statistically significant when p values <0.05.
Results
PHLDA3 overexpression was associated with poor prognosis of lung adenocarcinoma patients
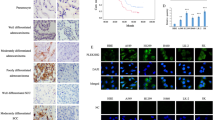
We examined the expression of PHLDA3 in 206 lung cancer and 108 corresponding normal lung tissue specimens using immunohistochemistry. PHLDA3 was primarily expressed in the cytoplasm. In corresponding normal lung tissues, positive expression of PHLDA3 was not observed in normal bronchial epithelial and alveolar cells, but was noted in 60 cases (29.1%) of NSCLC (Fig. 1A). The expression level of PHLDA3 was higher in lung cancer tissues than in normal lung tissues (p < 0.001) (Table 1).
A Immunohistochemical staining for PHLDA3 in normal lung tissues and lung cancer tissues. (magnification: ×100 or ×400). B Western blot analysis of PHLDA3 in lung cancer tissues and corresponding normal lung tissues. β-actin served as an internal control. N corresponding normal lung tissue, C lung cancer tissue. C Relative expression levels of PHLDA3 in lung cancer tissues and corresponding normal lung tissues (n = 60). **p < 0.01. D, E Kaplan–Meier plots of the overall survival of patients with lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LUSC) stratified by PHLDA3 expression, which were obtained from the Kaplan–Meier plotter database.
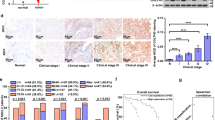
We also investigated the potential relationships between PHLDA3 expression and clinical parameters. As listed in Table 1, positive expression of PHLDA3 was significantly correlated with sex (p = 0.008), histological type (p = 0.009), TNM stage (p = 0.006), lymphatic metastasis (p = 0.049), and tumor T stage (p = 0.005) in lung cancers. The expression of PHLDA3 did not correlate with patient age (p = 0.160) or differentiation (p = 0.333) of lung malignancies. In LUAD, PHLDA3 expression was associated with TNM stage (p = 0.004), lymphatic metastasis (p = 0.032), and tumor T stage (p = 0.003), but not with patient age (p = 0.352), sex (p = 0.373), and differentiation (p = 0.185) (Table 2).
Western blot analysis confirmed that the expression levels of PHLDA3 were significantly higher in lung cancer tissues than in normal lung samples (n = 60; p < 0.01) (Fig. 1B, C). In addition, we also performed Kaplan–Meier survival analysis to determine whether PHLDA3 expression predicted patient outcomes, using the online Kaplan–Meier plotter database [27]. It was found that LUAD, but not LUSC, patients with high PHLDA3 expression had shorter overall survival (Fig. 1D, E).
PHLDA3 promoted lung cancer cell proliferation, invasion, and migration
To investigate the effects of PHLDA3 on the growth and mobility of lung cancer cells, we overexpressed PHLDA3 in H1299 and A549 cells via PHLDA3 gene transfection (Supplementary Fig. 1A), or downregulated PHLDA3 with siPHLDA3 (Supplementary Fig. 1B). Compared to that in the control cells, PHLDA3 overexpression was promoted, and PHLDA3 knockdown inhibited cell proliferation and colony formation (Fig. 2A–D). The results of Matrigel invasion and wound healing assays showed that PHLDA3 positively regulated the migratory and invasive phenotypes of lung cancer cells (Fig. 2E–H).
MTS assays (A, B) and colony formation assays (C, D) for H1299 and A549 cells transfected with PHLDA3 or siRNA against PHLDA3 (siPHLDA3). E, F Images and histograms of Matrigel invasion assays of the H1299 and A549 cells transfected with PHLDA3 or siPHLDA3. G, H Images and histograms of wound healing assays of the H1299 cells transfected with PHLDA3 or siPHLDA3. NC cells transfected with empty vector, siNC cells transfected with scrambled siRNA. *p < 0.05, **p < 0.01.
PHLDA3 activated the Wnt signaling pathway
After PHLDA3 gene transfection in H1299 and A549 cells, we evaluated the expression of key proteins involved in the Wnt signaling pathway. Compared with those in the control cells, the expression of active β-catenin, nuclear active β-catenin, total β-catenin, nuclear total β-catenin, and p-GSK3β was significantly increased, whereas that of GSK3β was decreased. Concomitantly, the expression levels of transcription factor TCF4 and target proteins of the Wnt signaling pathway, such as c-Myc, CyclinD1, and MMP7, were also increased (Fig. 3A, C). Conversely, knockdown of PHLDA3 decreased the expression levels of active β-catenin, nuclear active β-catenin, total β-catenin, nuclear total β-catenin, p-GSK3β, TCF4, c-Myc, cyclin D1, and MMP7, whereas GSK3β expression was increased in H1299 and A549 cells (Fig. 3B, D).
A, B Western blot analysis of active β-catenin, β-catenin, GSK3β, p-GSK3β, TCF4, c-Myc, CyclinD1, and MMP7 in the H1299 and A549 cells transfected with PHLDA3 or siRNA against PHLDA3 (siPHLDA3). GAPDH served as an internal control. C, D Western blot analysis for evaluating the nuclear and cytoplasmic levels of PHLDA3, active β-catenin, total β-catenin, ZEB1, and Twist in H1299 and A549 cells transfected with PHLDA3 or siPHLDA3. β-actin and LaminB served as internal cytoplasmic or nuclear controls, respectively. The relative protein levels in the control group were used to normalize protein levels in other groups. NC cells transfected with empty vector, siNC cells transfected with scrambled siRNA.
PHLDA3 facilitated the EMT process
Compared to that in the control cells, the expression of mesenchymal marker N-cadherin and the EMT-inducing transcription factors Snail, Twist, ZEB1, and MMP9, was significantly increased, whereas that of the epithelial markers E-cadherin, ZO1, and Claudin1 was decreased in the H1299 and A549 cells transfected with PHLDA3 (Fig. 4A). Immunofluorescent examination also confirmed that PHLDA3 upregulated the expression of mesenchymal markers N-cadherin and vimentin, whereas it downregulated the epithelial marker ZO1 (Fig. 4C). In contrast, the expression of N-cadherin, Snail, Twist, ZEB1, and MMP9 was decreased, whereas that of E-cadherin, ZO1, and Claudin1 was increased in siPHLDA3-treated H1299 and A549 cells (Fig. 4B). Immunofluorescent examination confirmed that the expression of N-cadherin and vimentin was decreased, and that of ZO1 was elevated in PHLDA3-depleted lung cancer cells (Fig. 4D).
A, B Western blot analysis of E-cadherin, ZO1, Claudin1, N-cadherin, Snail, Twist, ZEB1, and MMP9 in the H1299 and A549 cells transfected with PHLDA3 or siRNA against PHLDA3 (siPHLDA3). GAPDH served as an internal control. The relative protein levels in the control group were used to normalize protein levels in other groups. C, D Immunofluorescence staining of the indicated key EMT-related markers, N-cadherin, vimentin, and ZO1, in the H1299 cells transfected with PHLDA3, and the A549 cells transfected with siPHLDA3. NC cells transfected with empty vector, siNC cells transfected with scrambled siRNA.
PHLDA3 enhanced the proliferative and invasive abilities of lung cancer cells via the Wnt signaling pathway
We analyzed correlations between PHLDA3 and key proteins of the Wnt signaling pathway using the GEPIA database (http://gepia.cancer-pku.cn/), a newly developed web-based tool. PHLDA3 was found to be positively correlated with β-catenin (R = 0.18; p < 0.01) and negatively correlated with GSK3β (R = −0.12; p < 0.01) (Supplementary Fig. 2A, B). The expression levels of PHLDA3 and active β-catenin were also examined in eight pairs of lung cancer tissues and corresponding normal lung tissues. We found that PHLDA3 expression was positively correlated with active β-catenin expression (n = 8; R = 0.285; p < 0.05) (Supplementary Fig. 2C, D). To determine whether the functional activity of PHLDA3 is mediated through the Wnt signaling pathway, we treated H1299 and A549 cells with the Wnt inhibitor XAV-939. After ensuring a high transfection efficiency of PHLDA3 (MYC-tag), subsequent experiments revealed that XAV-939 reversed the upregulation of active β-catenin, p-GSK3β, c-Myc, Cyclin D1, and MMP7 induced by PHLDA3 overexpression while simultaneously reversing the downregulation of GSK3β (Fig. 5A). Induction of proliferation, colony formation, and Matrigel invasion of H1299 and A549 cells via PHLDA3 transfection was also partially reversed using XAV-939 (Fig. 5B–D).
A Western blot analysis of the expression levels of active β-catenin, GSK3β, p-GSK3β, c-Myc, Cyclin D1, and MMP7 in the H1299 and A549 cells with or without PHLDA3 transfection and XAV-939. GAPDH served as an internal control. The relative protein levels in the control group were used to normalize protein levels in other groups. B, C MTS assays and colony formation assays for the H1299 and A549 cells with or without PHLDA3 transfection and XAV-939. D Images and quantitation of Matrigel invasion assays of the H1299 and A549 cells with or without PHLDA3 transfection and XAV-939. Cells treated with DMSO served as the negative control. *p < 0.05, **p < 0.01.
PHLDA3 and GSK3β interactions modulated the Wnt signaling activity
As GSK3β is a key regulator of the Wnt signaling pathway, we verified the interaction between PHLDA3 and GSK3β in H1299 and A549 cells using co-immunoprecipitation assays (Fig. 6A). We then used the GSK3α/β inhibitor CHIR-99021 for further investigation. In H1299 and A549 cells, transfection with siPHLDA3 and treatment with CHIR-99021 reversed the inhibitory effects of PHLDA3 knockdown on cell proliferation, colony formation, and Matrigel invasion (Fig. 6B–D), indicating that the function of PHLDA3 in the Wnt signaling pathway and lung cancer cells partially depends on the regulation of GSK3β activity.
A Co-immunoprecipitation analysis of the interaction between PHLDA3 and GSK3β in H1299 and A549 cells. Cell lysates were immunoprecipitated with anti-MYC-tag antibody or control IgG, then examined for GSK3β expression using anti-GSK3β immunoblotting. B, C MTS assays and colony formation assays for the H1299 and A549 cells treated with siRNA against PHLDA3 (siPHLDA3) and/or GSK3α/β inhibitor CHIR-99021. D Images and quantitation of Matrigel invasion assays of the H1299 and A549 cells treated with PHLDA3 siRNA and/or GSK3α/β inhibitor CHIR-99021. Cells treated with DMSO served as the negative control. *p < 0.05, **p < 0.01.
Discussion
Previous studies examining PHLDA3 have focused on its effects on Akt activation via the inhibition of Akt binding to PI(3,4)P2 and PI(3,4,5)P3, which requires PHLDA3 to translocate to the plasma membrane [2, 8,9,10,11,12]. Studies exploring the role of PHLDA3 in tumors are limited. PHLDA3 has been identified as a tumor suppressor that suppresses Akt in neuroendocrine tumors [10,11,12]. However, Qiao et al. demonstrated that PHLDA3 inhibits somatic cell reprogramming by activating the Akt-GSK3β pathway [28]. In esophageal squamous cell carcinomas, low PHLDA3 expression is associated with postoperative tumor progression and recurrence, as well as poor patient prognosis [13], whereas PHLDA3 levels are significantly increased in head and neck squamous cell carcinoma [14]. Thus, it is possible that PHLDA3 exerts dual tumor suppressor and oncogenic roles based on the preferential mode of its regulatory effects on Akt downstream genes. The role and mechanism of PHLDA3 in lung cancer remain unclear. In the present study, we demonstrated that the expression of PHLDA3 was significantly higher in lung cancer tissues than in normal lung tissues. Moreover, the expression rate of PHLDA3 in LUAD was much higher than that in LUSCs. The high expression of PHLDA3 indicated advanced TNM stage and poor outcome in LUAD patients, but not in LUSC patients, which suggests that PHLDA3 plays a histology-specific role in lung cancers. This may be because LUAD and LUSC have different driver genes and signaling pathways. To the best of our knowledge, this is the first report on the expression, function, and clinical significance of PHLDA3 in lung adenocarcinoma.
Although the role of PHLDA3 has been reported in pancreatic neuroendocrine tumors, esophageal squamous cell carcinomas, and head and neck squamous cell carcinomas [11,12,13,14, 17], the mechanisms through which PHLDA3 promotes cancer progression remain unclear. To explore the mechanisms underlying the role of PHLDA3 in lung cancer cells, we confirmed that PHLDA3 promoted the proliferative and invasive abilities of lung cancer cells in vitro and investigated the regulatory effects of PHLDA3 on the Wnt signaling pathway and EMT, both of which play prominent roles in NSCLC [16, 20, 21]. PHLDA3 has been shown to be the target gene of p53; hence, we selected A549 (p53 wild type) and H1299 (p53 deletion type) cell lines to perform experiments in order to observe the effect of PHLDA3 on lung cancer cells with or without p53 [29]. The results obtained from the two cell lines were identical, indicating that p53 may not affect the function of PHLDA3 in lung cancer. We showed that PHLDA3 enhanced the expression and activation of β-catenin, and expression of the targets downstream of Wnt signaling, such as c-Myc, CyclinD1, and MMP7, as well as inhibited the expression and activation of GSK3β. PHLDA3 also regulated the expression of EMT-related proteins, confirming that PHLDA3 promotes EMT, a process involved early in the spread of cancer cells [30]. Furthermore, the Wnt inhibitor XAV-939, a potent tankyrase inhibitor that stimulates β-catenin degradation by stabilizing axin [31], partially reversed the PHLDA3-induced elevated expression of Wnt target genes and subsequent proteins, as well as modulated the proliferative and invasive capacities of lung cancer cells. Thus, we suggest that PHLDA3 facilitates the proliferation and invasion of lung cancer cells by promoting the Wnt signaling pathway.
How does PHLDA3 regulate the Wnt signaling pathway? The GEPIA database analysis suggested that PHLDA3 was positively correlated with β-catenin and negatively correlated with GSK3β. We also found, using co-immunoprecipitation assays, that PHLDA3 and GSK3β were associated with each other. We used the potent and selective GSK3α/β inhibitor CHIR-99021 for further investigation, as this compound is considered to be an activator of the Wnt signaling pathway [32,33,34]. CHIR-99021 reversed the inhibition of cell proliferation, colony formation, and Matrigel invasion induced by PHLDA3 knockdown. These results suggest that GSK3β is a regulatory target of PHLDA3; through PHLDA3, it led to the activation of the Wnt signaling pathway, as well as proliferation and invasion of lung cancer cells. However, the detailed mechanisms involved require further investigation.
In conclusion, PHLDA3 is highly expressed in lung cancers and is correlated with poor outcomes in patients with LUAD, making it a potential tumor malignancy marker and therapeutic target. PHLDA3 promotes the proliferation and invasion of lung cancer cells by regulating the activity of the Wnt signaling pathway and EMT.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Frank D, Mendelsohn CL, Ciccone E, Svensson K, Ohlsson R, Tycko B. A novel pleckstrin homology-related gene family defined by Ipl/Tssc3, TDAG51, and Tih1: tissue-specific expression, chromosomal location, and parental imprinting. Mamm Genome. 1999;10:1150–9.
Kawase T, Ohki R, Shibata T, Tsutsumi S, Kamimura N, Inazawa J, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–50.
Scheffzek K, Welti S. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 2012;586:2662–73.
Lee CG, Kang YJ, Kim HS, Moon A, Kim SG. Phlda3, a urine-detectable protein, causes p53 accumulation in renal tubular cells injured by cisplatin. Cell Biol Toxicol. 2015;31:121–30.
Han CY, Lim SW, Koo JH, Kim W, Kim SG. PHLDA3 overexpression in hepatocytes by endoplasmic reticulum stress via IRE1-Xbp1s pathway expedites liver injury. Gut. 2016;65:1377–88.
Sakata N, Yamaguchi Y, Chen Y, Shimoda M, Yoshimatsu G, Unno M, et al. Pleckstrin homology-like domain family A, member 3 (PHLDA3) deficiency improves islets engraftment through the suppression of hypoxic damage. PLoS ONE. 2017;12:e0187927.
Bensellam M, Chan JY, Lee K, Joglekar MV, Hardikar AA, Loudovaris T, et al. Phlda3 regulates beta cell survival during stress. Sci Rep. 2019;9:12827.
Liu J, Liu X, Hui X, Cai L, Li X, Yang Y, et al. Novel role for pleckstrin homology-like domain family a, member 3 in the regulation of pathological cardiac hypertrophy. J Am Heart Assoc. 2019;8:e011830.
Wang X, Li J, Yang Z, Wang L, Li L, Deng W, et al. phlda3 overexpression impairs specification of hemangioblasts and vascular development. Febs j. 2018;285:4071–81.
Chen Y, Ohki R. p53-PHLDA3-Akt network: the key regulators of neuroendocrine tumorigenesis. Int J Mol Sci. 2020;21:4098.
Ohki R, Saito K, Chen Y, Kawase T, Hiraoka N, Saigawa R, et al. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc Natl Acad Sci USA. 2014;111:E2404–13.
Takikawa M, Ohki R. A vicious partnership between AKT and PHLDA3 to facilitate neuroendocrine tumors. Cancer Sci. 2017;108:1101–8.
Muroi H, Nakajima M, Satomura H, Takahashi M, Yamaguchi S, Sasaki K, et al. Low PHLDA3 expression in oesophageal squamous cell carcinomas is associated with poor prognosis. Anticancer Res. 2015;35:949–54.
Saffarzadeh N, Ghafouri-Fard S, Rezaei Z, Aghazadeh K, Yazdani F, Mohebi M, et al. Expression Analysis of GRHL3 and PHLDA3 in Head and Neck Squamous Cell Carcinoma. Cancer Manag Res. 2020;12:4085–96.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Stewart DJ. Wnt signaling pathway in non-small cell lung cancer. J Natl Cancer Inst. 2014;106:djt356.
Aberle H, Bauer A, Stappert J, Kispert A, Kemler R. beta-catenin is a target for the ubiquitin-proteasome pathway. Embo J. 1997;16:3797–804.
Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–80.
Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205.
Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–96.
Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, et al. The 2015 World Health Organization Classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:1243–60.
Goldstraw P. Updated staging system for lung cancer. Surg Oncol Clin N Am. 2011;20:655–66.
Wang Y, Lei L, Zheng YW, Zhang L, Li ZH, Shen HY, et al. Odd-skipped related 1 inhibits lung cancer proliferation and invasion by reducing Wnt signaling through the suppression of SOX9 and β-catenin. Cancer Sci. 2018;109:1799–810.
Zheng YW, Li ZH, Lei L, Liu CC, Wang Z, Fei LR, et al. FAM83A promotes lung cancer progression by regulating the Wnt and Hippo signaling pathways and indicates poor prognosis. Front Oncol. 2020;10:180.
Fei LR, Huang WJ, Wang Y, Lei L, Li ZH, Zheng YW, et al. PRDM16 functions as a suppressor of lung adenocarcinoma metastasis. J Exp Clin Cancer Res. 2019;38:35.
Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8:e82241.
Qiao M, Wu M, Shi R, Hu W. PHLDA3 impedes somatic cell reprogramming by activating Akt-GSK3beta pathway. Sci Rep. 2017;7:2832.
Giaccone G, Battey J, Gazdar AF, Oie H, Draoui M, Moody TW. Neuromedin B is present in lung cancer cell lines. Cancer Res. 1992;52:2732s–2736s.
Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–30.
Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–20.
McManus EJ, Sakamoto K, Armit LJ, Ronaldson L, Shpiro N, Marquez R, et al. Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. Embo J. 2005;24:1571–83.
Naujok O, Lentes J, Diekmann U, Davenport C, Lenzen S. Cytotoxicity and activation of the Wnt/beta-catenin pathway in mouse embryonic stem cells treated with four GSK3 inhibitors. BMC Res Notes. 2014;7:273.
Ring DB, Johnson KW, Henriksen EJ, Nuss JM, Goff D, Kinnick TR, et al. Selective glycogen synthase kinase 3 inhibitors potentiate insulin activation of glucose transport and utilization in vitro and in vivo. Diabetes. 2003;52:588–95.
Funding
This study was supported by the Natural Science Foundation of Liaoning Province (Grant No. 2020-MS-179 to HTX).
Author information
Authors and Affiliations
Contributions
The authors contributed in the following way: LL conceived the study, conducted experiments, acquired and analyzed data, and wrote the manuscript; YW, ZHL, and ZFZ provided suggestions and participated in data analysis; WJH and YWZ contributed to the collection of the tissue specimens; CCL, ZW, and MQY contributed to data analysis; HTX was responsible for conception and supervision of the study, and wrote the manuscript. All authors have corrected the draft versions and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
This research was approved by the Human Research Ethics Committee of China Medical University, which is accredited by the National Council on Ethics in Human Research.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Lei, L., Wang, Y., Li, ZH. et al. PHLDA3 promotes lung adenocarcinoma cell proliferation and invasion via activation of the Wnt signaling pathway. Lab Invest 101, 1130–1141 (2021). https://doi.org/10.1038/s41374-021-00608-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-021-00608-3