Abstract
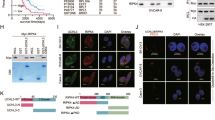
Melanoma-associated antigen A1 (MAGEA1) is member of the MAGE gene family that is expressed in male germ line cells and placenta under normal physiological conditions. Although MAGEA1’s expression levels have been evaluated as one of the cancer testis (CT) antigens for immunotherapy in melanoma and several other cancers, its functional role and signaling mechanisms are largely unknown. In this study, we examined the functional involvement and signaling mechanisms of MAGEA1 in breast and ovarian cancer cells. Inhibitory effects of MAGEA1 on cell proliferation and migration were observed using both gain-of- (MAGEA1 overexpression) and loss-of- (siRNA interference) function approaches in breast (MCF-7 and MDA-MB-231) and ovarian (SKOV3 and SKOV3ip) cancer cell lines. We revealed a novel interaction between MAGEA1 and the intracellular segment of NOTCH1 receptor (NICD1). MAGEA1 reduced NICD1’s stability by promoting the ubiquitin modification of NICD1. MAGEA1 also interacted with FBXW7, subunit of E3 ubiquitin protein ligase complex SCF, and the latter was functionally involved in NICD1 ubiquitination and degradation. In addition, siRNA interference of FBXW7 reversed the inhibitory effect of MAGEA1 on migration and proliferation of MCF-7 and MDA-MB-231 cells. These newly discovered MAGEA1–NICD1 and MAGEA1–FBXW7 interactions have potential clinical implications in breast and ovarian cancer treatment.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Traversari C, van der Bruggen P, Van den Eynde B, Hainaut P, Lemoine C, Ohta N et al. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics 1992; 35: 145–152.
Vanderbruggen P, Traversari C, Chomez P, Lurquin C, Deplaen E, Vandeneynde B et al. A gene encoding an antigen recognized by cytolytic lymphocytes-T on a human-melanoma. Science 1991; 254: 1643–1647.
Ladelfa MF, Peche LY, Toledo MF, Laiseca JE, Schneider C, Monte M . Tumor-specific MAGE proteins as regulators of p53 function. Cancer Lett 2012; 325: 11–17.
Kloudova K, Hromadkova H, Partlova S, Brtnicky T, Rob L, Bartunkova J et al. Expression of tumor antigens on primary ovarian cancer cells compared to established ovarian cancer cell lines. Oncotarget 2016; 7: 46120–46126.
Otte M, Zafrakas M, Riethdorf L, Pichlmeier U, Loning T, Janicke F et al. MAGE-A gene expression pattern in primary breast cancer. Cancer Res 2001; 61: 6682–6687.
Matkovic B, Juretic A, Spagnoli GC, Separovic V, Gamulin M, Separovic R et al. Expression of MAGE-A and NY-ESO-1 cancer/testis antigens in medullary breast cancer: retrospective immunohistochemical study. Croat Med J 2011; 52: 171–177.
Abd-Elsalam EA, Ismaeil NA . Melanoma-associated antigen genes: a new trend to predict the prognosis of breast cancer patients. Med Oncol 2014; 31: 285.
Larsen RD, Rajan VP, Ruff MM, Kukowska-Latallo J, Cummings RD, Lowe JB . Isolation of a cDNA encoding a murine UDPgalactose:beta-D-galactosyl- 1,4-N-acetyl-D-glucosaminide alpha-1,3-galactosyltransferase: expression cloning by gene transfer. Proc Natl Acad Sci USA 1989; 86: 8227–8231.
Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615.
Acar A, Simoes BM, Clarke RB, Brennan K . A role for notch signalling in breast cancer and endocrine resistance. Stem Cells Int 2016; 2016: 2498764.
Bolos V, Mira E, Martinez-Poveda B, Luxan G, Canamero M, Martinez C et al. Notch activation stimulates migration of breast cancer cells and promotes tumor growth. Breast Cancer Res 2013; 15: 15.
Nickoloff BJ, Osborne BA, Miele L . Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene 2003; 22: 6598–6608.
Stylianou S, Clarke RB, Brennan K . Aberrant activation of Notch signaling in human breast cancer. Cancer Res 2006; 66: 1517–1525.
Bray SJ . Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 2006; 7: 678–689.
Fortini ME . Notch signaling: the core pathway and its posttranslational regulation. Dev Cell 2009; 16: 633–647.
Bao B, Wang ZW, Ali S, Kong DJ, Li YW, Ahmad A et al. Notch-1 induces epithelial-mesenchymal transition consistent with cancer stem cell phenotype in pancreatic cancer cells. Cancer Lett 2011; 307: 26–36.
Hu YY, Zheng MH, Zhang R, Liang YM, Han H . Notch signaling pathway and cancer metastasis. Adv Exp Med Biol 2012; 727: 186–198.
Saha SK, Choi HY, Kim BW, Dayem AA, Yang GM, Kim KS et al. KRT19 directly interacts with beta-catenin/RAC1 complex to regulate NUMB-dependent NOTCH signaling pathway and breast cancer properties. Oncogene 2017; 36: 332–349.
Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem 2001; 276: 34371–34378.
Pui CH . T cell acute lymphoblastic leukemia: NOTCHing the way toward a better treatment outcome. Cancer Cell 2009; 15: 85–87.
Wu GY, Lyapina S, Das I, Li JH, Gurney M, Pauley A et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol Cell Biol 2001; 21: 7403–7415.
Laduron S, Deplus R, Zhou S, Kholmanskikh O, Godelaine D, De Smet C et al. MAGE-A1 interacts with adaptor SKIP and the deacetylase HDAC1 to repress transcription. Nucleic Acids Res 2004; 32: 4340–4350.
Zhou SF, Fujimuro M, Hsieh JJD, Chen L, Miyamoto A, Weinmaster G et al. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol 2000; 20: 2400–2410.
De Plaen E, De Backer O, Arnaud D, Bonjean B, Chomez P, Martelange V et al. A new family of mouse genes homologous to the human MAGE genes. Genomics 1999; 55: 176–184.
Gillespie AM, Coleman RE . The potential of melanoma antigen expression in cancer therapy. Cancer Treat Rev 1999; 25: 219–227.
Friedl P, Wolf K . Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003; 3: 362–374.
Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011; 474: 609–615.
Thiery JP, Acloque H, Huang RYJ, Nieto MA . Epithelial-mesenchymal transitions in development and disease. Cell 2009; 139: 871–890.
Singh A, Settleman J . EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010; 29: 4741–4751.
Hochstrasser M . Ubiquitin-dependent protein degradation. Annu Rev Genet 1996; 30: 405–439.
Pickart CM . Mechanisms underlying ubiquitination. Annu Rev Biochem 2001; 70: 503–533.
Doyle JM, Gao J, Wang J, Yang M, Potts PR . MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell 2010; 39: 963–974.
Hao JQ, Song X, Wang JJ, Guo CL, Li Y, Li B et al. Cancer-testis antigen MAGE-C2 binds Rbx1 and inhibits ubiquitin ligase-mediated turnover of cyclin E. Oncotarget 2015; 6: 42028–42039.
Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol 2015; 12: 445–464.
Shukla G, Khera HK, Srivastava AK, Khare P, Patidar R, Saxena R . Therapeutic potential, challenges and future perspective of cancer stem cells in translational oncology: a critical review. Curr Stem Cell Res Ther 2017; 12: 207–224.
Brzozowa-Zasada M, Piecuch A, Dittfeld A, Mielanczyk L, Michalski M, Wyrobiec G et al. Notch signalling pathway as an oncogenic factor involved in cancer development. Contemp Oncol (Pozn) 2016; 20: 267–272.
Weon JL, Potts PR . The MAGE protein family and cancer. Curr Opin Cell Biol 2015; 37: 1–8.
Zhang S, Zhou X, Yu H, Yu Y . Expression of tumor-specific antigen MAGE, GAGE and BAGE in ovarian cancer tissues and cell lines. BMC Cancer 2010; 10: 163.
Daudi S, Eng KH, Mhawech-Fauceglia P, Morrison C, Miliotto A, Beck A et al. Expression and immune responses to MAGE antigens predict survival in epithelial ovarian cancer. PloS One 2014; 9: e104099.
Wang D, Wang JY, Ding N, Li YJ, Yang YR, Fang XD et al. MAGE-A1 promotes melanoma proliferation and migration through C-JUN activation. Biochem Biophys Res Commun 2016; 473: 959–965.
Marcar L, Ihrig B, Hourihan J, Bray SE, Quinlan PR, Jordan LB et al. MAGE-A cancer/testis antigens inhibit MDM2 ubiquitylation function and promote increased levels of MDM4. PloS One 2015; 10: e0127713.
Askew EB, Bai SX, Hnat AT, Minges JT, Wilson EM . Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators. J Biol Chem 2009; 284: 34793–34808.
Marcar L, Maclaine NJ, Hupp TR, Meek DW . Mage-A cancer/testis antigens inhibit p53 function by blocking its interaction with chromatin. Cancer Res 2010; 70: 10362–10370.
Monte M, Simonatto M, Peche LY, Bublik DR, Gobessi S, Pierotti MA et al. MAGE-A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci USA 2006; 103: 11160–11165.
Peche LY, Scolz M, Ladelfa MF, Monte M, Schneider C . MageA2 restrains cellular senescence by targeting the function of PMLIV/p53 axis at the PML-NBs. Cell Death Differ 2012; 19: 926–936.
Tseng HY, Chen LH, Ye Y, Tay KH, Jiang CC, Guo ST et al. The melanoma-associated antigen MAGE-D2 suppresses TRAIL receptor 2 and protects against TRAIL-induced apoptosis in human melanoma cells. Carcinogenesis 2012; 33: 1871–1881.
Potapova O, Basu S, Mercola D, Holbrook NJ . Protective role for c-Jun in the cellular response to DNA damage. J Biol Chem 2001; 276: 28546–28553.
She QB, Chen N, Bode AM, Flavell RA, Dong Z . Deficiency of c-Jun-NH(2)-terminal kinase-1 in mice enhances skin tumor development by 12-O-tetradecanoylphorbol-13-acetate. Cancer Res 2002; 62: 1343–1348.
Uhlirova M, Jasper H, Bohmann D . Non-cell-autonomous induction of tissue overgrowth by JNK/Ras cooperation in a Drosophila tumor model. Proc Natl Acad Sci USA 2005; 102: 13123–13128.
Kim HL, Vander Griend DJ, Yang X, Benson DA, Dubauskas Z, Yoshida BA et al. Mitogen-activated protein kinase kinase 4 metastasis suppressor gene expression is inversely related to histological pattern in advancing human prostatic cancers. Cancer Res 2001; 61: 2833–2837.
Heasley LE, Han SY . JNK regulation of oncogenesis. Mol Cells 2006; 21: 167–173.
Kitade S, Onoyama I, Kobayashi H, Yagi H, Yoshida S, Kato M et al. FBXW7 is involved in the acquisition of the malignant phenotype in epithelial ovarian tumors. Cancer Sci 2016; 107: 1399–1405.
Weinberger M, Tierney WM, Booher P, Katz BP . Can the provision of information to patients with osteoarthritis improve functional status? A randomized, controlled trial. Arthritis Rheum 1989; 32: 1577–1583.
Kozakova L, Vondrova L, Stejskal K, Charalabous P, Kolesar P, Lehmann AR et al. The melanoma-associated antigen 1 (MAGEA1) protein stimulates the E3 ubiquitin-ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex. Cell Cycle 2015; 14: 920–930.
Minges JT, Su SF, Grossman G, Blackwelder AJ, Pop EA, Mohler JL et al. Melanoma antigen-A11 (MAGE-A11) enhances transcriptional activity by linking androgen receptor dimers. J Biol Chem 2013; 288: 1939–1952.
Acknowledgements
This work was supported in part by The National Basic Research Program of China (Grant no. 2007CB914401) and the National High Technology Research and Development Program of China (Grant no. 2006AA02Z4A6) to JS; as well as in part by the Mary Fendrich-Hulman Charitable Trust Fund to YX. We would like to thank Kevin McClelland for editing the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhao, J., Wang, Y., Mu, C. et al. MAGEA1 interacts with FBXW7 and regulates ubiquitin ligase-mediated turnover of NICD1 in breast and ovarian cancer cells. Oncogene 36, 5023–5034 (2017). https://doi.org/10.1038/onc.2017.131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.131
This article is cited by
-
FBXW7 in breast cancer: mechanism of action and therapeutic potential
Journal of Experimental & Clinical Cancer Research (2023)
-
Clinical significance of FBXW7 loss of function in human cancers
Molecular Cancer (2022)
-
VIVA1: a more invasive subclone of MDA-MB-134VI invasive lobular carcinoma cells with increased metastatic potential in xenograft models
British Journal of Cancer (2022)
-
FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2
Molecular Cancer (2021)
-
Downregulation of the ubiquitin ligase KBTBD8 prevented epithelial ovarian cancer progression
Molecular Medicine (2020)