Abstract
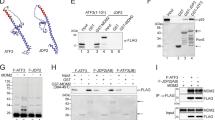
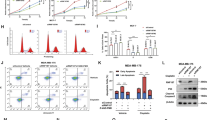
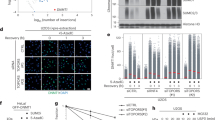
Ubiquitin linkage is critical in directing the cellular fate of a ubiquitinated protein. Although K48-linked polyubiquitination of p53 leads to its degradation, whether K48-independent ubiquitin linkages are involved in p53 activation remains unknown. Here, we show that FATS acts as a p53 activator by inhibiting Mdm2 binding to p53 and stimulating non-proteolytic polyubiquitination of p53. Knockdown of FATS impairs p53 stabilization and activation in response to DNA damage. Furthermore, the NH2-terminal domain of FATS is sufficient to exhibit ubiquitin ligase (E3) activity and assemble ubiquitin polymers through K11-, K29- and K63-linkages, independently of the ubiquitin-conjugating enzyme (E2). FATS promotes p53-dependent transcription of p21, leading to robust checkpoint response. The E3 activity of FATS is required for promoting p53 stability and activation in response to DNA damage. Our findings reveal K48-linkage-independent non-linear polyubiquitination of p53 as a new barcode for p53 activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Rotin D, Kumar S . Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 2009; 10: 398–409.
Kerscher O, Felberbaum R, Hochstrasser M . Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 2006; 22: 159–180.
Mukhopadhyay D, Riezman H . Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 2007; 315: 201–205.
Ulrich HD, Walden H . Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Boil 2010; 11: 479–489.
Pickart CM, Fushman D . Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 2004; 8: 610–616.
Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 2009; 137: 133–145.
Lee JT, Gu W . The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 2010; 17: 86–92.
Jones SN, Roe AE, Donehower LA, Bradley A . Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 1995; 378: 206–208.
Montes de Oca Luna R, Wagner DS, Lozano G . Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 1995; 378: 203–206.
Haupt Y, Maya R, Kazaz A, Oren M . Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–299.
Shieh SY, Ikeda M, Taya Y, Prives C . DNA damage-induced phosphorylation of p53 alleviates inhibition by Mdm2. Cell 1997; 91: 325–334.
Ito A, Kawaguchi Y, Lai CH, Kovacs JJ, Higashimoto Y, Appella E et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J 2002; 21: 6236–6245.
Tang Y, Zhao W, Chen Y, Zhao Y, Gu W . Acetylation is indispensable for p53 activation. Cell 2008; 133: 612–626.
Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429: 86–92.
Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 2003; 112: 779–791.
Wu X, Bayle JH, Olson D, Levine AJ . The p53-Mdm2 autoregulatory feedback loop. Genes Dev 1993; 7: 1126–1132.
Jain AK, Barton MC . Making sense of ubiquitin ligases that regulate p53. Cancer Biol Ther 2010; 10: 665–672.
Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E et al. COP1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest 2011; 121: 1329–1343.
Jung YS, Hakem A, Hakem R, Chen X . Pirh2 E3 ubiquitin ligase monoubiquitinates DNA polymerase eta to suppress translesion DNA synthesis. Mol Cell Biol 2011; 31: 3997–4006.
Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell 2006; 127: 775–788.
Durkin SG, Glover TW . Chromosome fragile sites. Annu Rev Genet 2007; 41: 169–192.
Ma K, Qiu L, Mrasek K, Zhang J, Liehr T, Quintana LG et al. Common fragile sites: genomic hotspots of DNA damage and carcinogenesis. Int J Mol Sci 2012; 13: 11974–11999.
Li Z, Zhang Q, Mao JH, Weise A, Mrasek K, Fan X et al. An HDAC1-binding domain within FATS bridges p21 turnover to radiation-induced tumorigenesis. Oncogene 2010; 29: 2659–2671.
Zhang X, Zhang Q, Zhang J, Qiu L, Yan S, Feng J et al. FATS is a transcriptional target of p53 and associated with antitumor activity. Mol Cancer 2010; 9: 244.
Zhang J, Gu L, Zhao L, Zhang X, Qiu L, Li Z . Expression level of novel tumor suppressor gene FATS is associated with the outcome of node positive breast cancer. Chin Med J 2011; 124: 2894–2898.
Tian Y, Zhang J, Yan S, Qiu L, Li Z . FATS expression is associated with cisplatin sensitivity in non small cell lung cancer. Lung Cancer 2012; 76: 416–422.
Mao JH, Li J, Jiang T, Li Q, Wu D, Perez-Losada J et al. Genomic instability in radiation-induced mouse lymphoma from p53 heterozygous mice. Oncogene 2005; 24: 7924–7934.
Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC et al. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem 2004; 279: 42169–42181.
Wenzel DM, Lissounov A, Brzovic PS, Klevit RE . UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 2011; 474: 105–108.
Laine A, Ronai Z . Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene 2007; 26: 1477–1483.
Laine A, Topisirovic I, Zhai D, Reed JC, Borden KL, Ronai Z . Regulation of p53 localization and activity by Ubc13. Mol Cell Biol 2006; 26: 8901–8913.
Vousden KH, Prives C . Blinded by the light: the growing complexity of p53. Cell 2009; 137: 413–431.
Ye Y, Rape M . Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 2009; 10: 755–764.
Hurley JH, Lee S, Prag G . Ubiquitin-binding domains. Biochem J 2006; 399: 361–372.
Bellail AC, Olson JJ, Yang X, Chen ZJ, Hao C . A20 ubiquitin ligase-mediated polyubiquitination of RIP1 inhibits caspase-8 cleavage and TRAIL-mediated apoptosis in glioblastoma. Cancer Discov 2012; 2: 140–155.
Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit REA . UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 2006; 21: 873–880.
Wu H, Pomeroy SL, Ferreira M, Teider N, Mariani J, Nakayama KI et al. UBE4B promotes Hdm2-mediated degradation of the tumor suppressor p53. Nat Med 2011; 17: 347–355.
Najafi SM, Li Z, Makino K, Shao R, Hung MC . The adenoviral E1A induces p21WAF1/CIP1 expression in cancer cells. Biochem Biophys Res Commun 2003; 305: 1099–1104.
Li Z, Day CP, Yang JY, Tsai WB, Lozano G, Shih HM et al. Adenoviral E1A targets Mdm4 to stabilize tumor suppressor p53. Cancer Res 2004; 64: 9080–9085.
Acknowledgements
We are grateful to Dr X Lu (The University of Texas MD Anderson Cancer Center, USA) for providing us with HA-Mdm2 plasmid. This work was supported by grants from Ministry of Science and Technology of China 973-program concept award (2009CB526407 to ZL), National Natural Science Foundation of China (81272283 to ZL), and Tianjin Municipal Science and Technology Foundation (10JCZDJC18600 to ZL).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Yan, S., Qiu, L., Ma, K. et al. FATS is an E2-independent ubiquitin ligase that stabilizes p53 and promotes its activation in response to DNA damage. Oncogene 33, 5424–5433 (2014). https://doi.org/10.1038/onc.2013.494
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2013.494
Keywords
This article is cited by
-
Gene losses may contribute to subterranean adaptations in naked mole-rat and blind mole-rat
BMC Biology (2022)
-
Loss of fragile site-associated tumor suppressor promotes antitumor immunity via macrophage polarization
Nature Communications (2021)
-
FATS regulates polyamine biosynthesis by promoting ODC degradation in an ERβ-dependent manner in non-small-cell lung cancer
Cell Death & Disease (2020)
-
DNA methylation modifier LSH inhibits p53 ubiquitination and transactivates p53 to promote lipid metabolism
Epigenetics & Chromatin (2019)
-
ALMS1 and Alström syndrome: a recessive form of metabolic, neurosensory and cardiac deficits
Journal of Molecular Medicine (2019)