Abstract
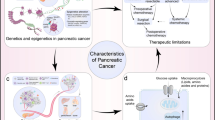
Current standard therapies for pancreatic ductal adenocarcinoma have failed to attenuate the aggressiveness of this disease or confer notable improvements in survival. Previous molecular research into pancreatic cancers, along with advances in sequencing technologies, have identified many altered genes in patients with pancreatic cancer and revealed the marked genetic heterogeneity of individual tumors. Thus, the lack of success of conventional empiric therapy can be partly attributed to the underlying heterogeneity of pancreatic tumors. The genetic alterations that have been detected in pancreatic cancer range from simple mutations at the level of base pairs to complex chromosomal structural changes and rearrangements. The identification of molecular changes that are unique to an individual patient's tumors, and the subsequent development of strategies to target the tumors in a personalized approach to therapeutics, is a necessary advance to improve therapy for patients with this disease.
Key Points
-
Pancreatic cancer is a lethal malignancy; the treatment of patients with this disease is a therapeutic challenge
-
Current therapies confer only modest increases in survival, often as little as a few months
-
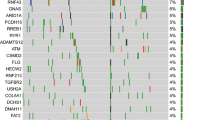
Molecular and genomic research into pancreatic cancer has demonstrated marked heterogeneity among tumors that might underlie differing responses to standard therapy
-
Improved therapeutic outcomes might be realized in some patients through a personalized approach to targeting patient-specific genetic alterations in tumors
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Yeo, T. P. et al. Pancreatic cancer. Curr. Probl. Cancer 26, 176–275 (2002).
Jemal, A., Seigel, R., Xu, J. & Ward, E. Cancer statistics, 2010. CA Cancer J. Clin. 60, 277–300 (2010).
Hezel, A. F., Kimmelman, A. C., Stanger, B. Z., Bardeesy, N. & DePinho, R. A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 20, 1218–1249 (2006).
Feldmann, G. & Maitra, A. Molecular genetics of pancreatic ductal adenocarcinomas and recent implications for translational efforts. J. Mol. Diagn. 10, 111–122 (2008).
Gräntzdörffer, I., Carl-McGrath, S., Ebert, M. P. & Röcken, C. Proteomics of pancreatic cancer. Pancreas 36, 329–336 (2008).
Li, D., Xie, K., Wolff, R. & Abbruzzese, J. L. Pancreatic cancer. Lancet 363, 1049–1057 (2004).
Klöppel, G., Solcia, E., Longnecker, D. S., Capella, C. & Sobin, L. H. in International Histological Classification of Tumours 2nd edn 1–61 (Springer/World Health Organization, Berlin, 1998).
Hruban, R. H., Goggins, M., Parsons, J. & Kern, S. E. Progression model for pancreatic cancer. Clin. Cancer Res. 6, 2969–2972 (2000).
Buchholz, M. et al. Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene 24, 6626–6636 (2005).
Olive, K. P. et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324, 1457–1461 (2009).
Chu, G. C., Kimmelman, A. C., Hezel, A. F. & DePinho, R. A. Stromal biology of pancreatic cancer. J. Cell. Biochem. 101, 887–907 (2007).
Neesse, A. et al. Stromal biology and therapy in pancreatic cancer. Gut 60, 861–868 (2011).
Wang, Z. et al. Pancreatic cancer: understanding and overcoming chemoresistance. Nat. Rev. Gastroenterol. Hepatol. 8, 27–33 (2011).
Wang, Z. et al. Acquisition of epithelial-mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Res. 69, 2400–2407 (2009).
Shah, A. N. et al. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann. Surg. Oncol. 14, 3629–3637 (2007).
Hermann, P. C. et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 (2007).
Hong, S. P., Wen, J., Bang, S., Park, S. & Song, S. Y. CD44-positive cells are responsible for gemcitabine resistance in pancreatic cancer cells. Int. J. Cancer 125, 2323–2331 (2009).
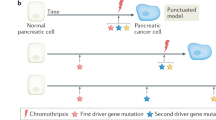
Campbell, P. J. et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467, 1109–1113 (2010).
Biankin, A. V. & Hudson, T. J. Somatic variation and cancer: therapies lost in the mix. Hum. Genet. 130, 79–91 (2011).
Hong, S. M., Park, J. Y., Hruban, R. H. & Goggins, M. Molecular signatures of pancreatic cancer. Arch. Pathol. Lab. Med. 135, 716–727 (2011).
Harris, T. J. & McCormick, F. The molecular pathology of cancer. Nat. Rev. Clin. Oncol. 7, 251–265 (2010).
Lynch, H. T. et al. Familial pancreatic cancer: a review. Semin. Oncol. 23, 251–275 (1996).
Bignell, G. R. et al. Signatures of mutation and selection in the cancer genome. Nature 463, 893–898 (2010).
Bardeesy, N. & DePinho, R. A. Pancreatic cancer biology and genetics. Nat. Rev. Cancer 2, 897–909 (2002).
Hansel, D. E., Kern, S. E. & Hruban, R. H. Molecular pathogenesis of pancreatic cancer. Annu. Rev. Genomics Hum. Genet. 4, 237–256 (2003).
Baumgart, M., Heinmöller, E., Horstmann, O., Becker, H. & Ghadimi, B. M. The genetic basis of sporadic pancreatic cancer. Cell. Oncol. 27, 3–13 (2005).
Wong, H. H. & Lemoine, N. R. Pancreatic cancer: molecular pathogenesis and new therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 6, 412–422 (2009).
Klimstra, D. S. & Longnecker, D. S. K-ras mutations in pancreatic ductal proliferative lesions. Am. J. Pathol. 145, 1547–1550 (1994).
Rozenblum, E. et al. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 57, 1731–1734 (1997).
Rachagani, S. et al. Activated KrasG12D is associated with invasion and metastasis of pancreatic cancer cells through inhibition of E-cadherin. Br. J. Cancer 104, 1038–1048 (2011).
Pugliese, V. et al. Pancreatic intraductal sampling during ERCP in patients with chronic pancreatitis and pancreatic cancer: cytologic studies and k-ras-2 codon 12 molecular analysis in 47 cases. Gastrointest. Endosc. 54, 595–599 (2001).
Van Laethem, J. L. et al. Relative contribution of Ki-ras gene analysis and brush cytology during ERCP for the diagnosis of biliary and pancreatic diseases. Gastrointest. Endosc. 47, 479–485 (1998).
Luttges, J. et al. The K-ras mutation pattern in pancreatic ductal adenocarcinoma usually is identical to that in associated normal, hyperplastic, and metaplastic ductal epithelium. Cancer 85, 1703–1710 (1999).
Hruban, R. H., Wilentz, R. & Kern, S. E. Genetic progression in the pancreatic ducts. Am. J. Pathol. 156, 1821–1825 (2000).
Calhoun, E. S. et al. BRAF and FBXW7 (CDC4, FBW7, AGO, SEL10) mutations in distinct subsets of pancreatic cancer: potential therapeutic targets. Am. J. Pathol. 163, 1255–1260 (2003).
Cheng, J. Q. et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl Acad. Sci. USA 93, 3636–3641 (1996).
Ruggeri, B. A., Huang, L., Wood, M., Cheng, J. Q. & Testa, J. R. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol. Carcinog. 21, 81–86 (1998).
Wallrapp, C. et al. Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res. 57, 3135–3139 (1997).
Caldas, C. et al. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat. Genet. 8, 27–32 (1994).
Ruggeri, B. A. et al. Human pancreatic carcinomas and cell lines reveal frequent and multiple alterations in the p53 and Rb-1 tumor-suppressor genes. Oncogene 7, 1503–1511 (1992).
Scarpa, A. et al. Pancreatic adenocarcinomas frequently show p53 gene mutations. Am. J. Pathol. 142, 1534–1543 (1993).
Hahn, S. A. et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271, 350–353 (1996).
Vogelstein, B. & Kinzler, K. W. Cancer genes and the pathways they control. Nat. Med. 10, 789–799 (2004).
Maitra, A. & Hruban, R. H. Pancreatic cancer. Annu. Rev. Pathol. 3, 157–188 (2008).
Wu, X. et al. HOXB7 homeodomain protein is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 66, 9527–9534 (2006).
Chen, H. et al. Hoxb7 inhibits transgenic HER-2/neu-induced mouse mammary tumor onset but promotes progression and lung metastasis. Cancer Res. 68, 3637–3644 (2008).
De Souza Setubal Destro, M. F. et al. Overexpression of HOXB7 homebox gene in oral cancer induces ceullar proliferation and is associated with poor prognosis. Int. J. Oncol. 36, 141–149 (2010).
Kim, Y. S., Gum, J. R. Jr, Crawley, S. C., Deng G. & Ho, J. J. Mucin gene and antigen expression in biliopancreatic carcinogenesis. Ann. Oncol. 10 (Suppl. 4), 51–55 (1999).
Kim, G. E. et al. Aberrant expression of MUC5AC and MUC6 gastric mucins and sialyl Tn antigen in intraepithelial neoplasms of the pancreas. Gastroenterology 123, 1052–1060 (2002).
Monges, G. M. et al. Differential MUC1 expression in normal and neoplastic human pancreatic tissue. An immunohistochemical study of 60 samples. Am. J. Clin. Pathol. 112, 635–640 (1999).
Besmer, D. M. et al. Pancreatic ductal adenocarcinoma (PDA) mice lacking mucin 1 have a profound defect in tumor growth and metastasis. Cancer Res. 71, 4434–4442 (2011).
Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Ding, L. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 455, 1069–1075 (2008).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Varela, I. et al. Exome sequencing identifies frequent mutations of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Jones, S. et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Wood, L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007).
Morris, J. P. 4th, Wang, S. C. & Hebrok, M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer 10, 683–695 (2010).
Stuart, D. & Sellers, W. R. Linking somatic genetic alterations to cancer therapeutics. Curr. Opin. Cell Biol. 21, 304–310 (2009).
International Cancer Genome Consortium. International network of cancer genome projects. Nature 464, 993–998 (2010).
Suzuki, A. et al. Identification of SMURF1 as a possible target for 7q21.3–221 amplification detected in a pancreatic cancer cell line by in-house array-based comparative genomic hybridization. Cancer Sci. 99, 986–994 (2008).
Fu, B., Luo, M., Lakkur, S., Lucito, R. & Iacobuzio-Donahue, C. A. Frequent genomic copy number gain and overexpression of GATA-6 in pancreatic carcinoma. Cancer Biol. Ther. 7, 1593–1601 (2008).
Heidenblad, M. et al. Genome-wide array-based comparative genomic hybridization reveals multiple amplification targets and novel homozygous deletions in pancreatic carcinoma cell lines. Cancer Res. 64, 3052–3059 (2004).
Holzmann, K. et al. Genomic DNA-chip hybridization reveals a higher incidence of genomic amplifications in pancreatic cancer than conventional comparative genomic hybridization and leads to the identification of novel candidate genes. Cancer Res. 64, 4428–4433 (2004).
Kitoh, H. et al. Comparative genomic hybridization analysis for pancreatic cancer specimens obtained by endoscopic ultrasonography-guided fine-needle aspiration. J. Gastroenterol. 40, 511–517 (2005).
Loukopoulous, P. et al. Genome-wide array-based comparative genomic hybridization analysis of pancreatic adenocarcinome: identification of genetic indicators that predict patient outcome. Cancer Sci. 98, 392–400 (2007).
Zhong, Y. et al. GATA6 activates Wnt signaling in pancreatic cancer by negatively regulating the Wnt antagonist Dickkopf-1. PLoS ONE 6, e22129 (2011).
Genomic Health. Oncotype DX [online], (2010).
Collisson, E. A. et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 17, 500–503 (2011).
Chelala, C. et al. Pancreatic expression database: a generic model for the organization, integration and mining of complex cancer datasets. BMC Genomics 8, 439 (2007).
International Cancer Genome Consortium Data Coordination Center. Data Portal [online], (2011).
Vincent, A. et al. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin. Cancer Res. 17, 4341–4354 (2011).
Bloomston, M. et al. MircoRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297, 1901–1908 (2007).
Dillhoff, M., Lieu, J., Frankel, W., Croce, C. & Bloomston, M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J. Gastrointest. Surg. 12, 2171–2176 (2008).
Lee, E. J. et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int. J. Cancer 120, 1046–1054 (2007).
Szafranska, A. E. et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 26, 4442–4452 (2007).
Zhang, Y. et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J. Surg. 33, 698–709 (2009).
Kent, O. A. et al. A resource for analysis of microRNA expression and function in pancreatic ductal adenocarcinoma cells. Cancer Biol. Ther. 8, 2013–2024 (2009).
Burris, H. A. 3rd et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J. Clin. Oncol. 15, 2403–2413 (1997).
Moore, M. J. et al. Erlotinib plus gemcitabine compared with gemcitabine alone in patients with advanced pancreatic cancer: a Phase III trial of the National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 25, 1960–1966 (2007).
Conroy, T. et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 364, 1817–1825 (2011).
Stratton, M. R. Exploring the genomes of cancer cells: progress and promise. Science 33, 1553–1558 (2011).
Esteva, F. J., Yu, D., Hung., M. C. & Horobagyi, G. N. Molecular predictors of response to trastuzumab and lapatinib in breast cancer. Nat. Rev. Clin. Oncol. 7, 98–107 (2010).
Genentech. FDA Approves Zelboraf (Vemurafenib) and Companion Diagnostic for BRAF Mutation-Positive Metastatic Melanoma, a Deadly Form of Skin Cancer [online], (2011).
Villarroel, M. C. et al. Personalizing cancer treatment in the age of global genomic analyses: PALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol. Cancer Ther. 10, 3–8 (2011).
Xia, B. et al. Control of BRAC2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 (2006).
Jones, S. et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324, 217 (2009).
Couch, F. J. et al. The prevalence of BRCA2 mutations in familial pancreatic cancer. Cancer Epidemiol. Biomarkers Prev. 16, 342–346 (2007).
Iacobuzio-Donahue, C. A. et al. DPC4 gene status of the primary carcinoma correlates with patterns of failure in patients with pancreatic cancer. J. Clin. Oncol. 27, 1806–1813 (2009).
Armstrong, M. D. et al. An effective personalized approach to a rare tumor: prolonged survival in metastatic pancreatic acinar cell carcinoma based on genetic analysis and cell line development. J. Cancer 2, 142–152 (2011).
Solinas-Toldo, S. et al. Mapping of chromosomal imbalances in pancreatic carcinoma by comparative genomic hybridization. Cancer Res. 56, 3803–3807 (1996).
Mahlamäki, E. H. et al. Comparative genomic hybridization reveals frequent gains of 20q, 8q, 11q, 12p, and 17q and losses of 18q, 9p and 15q in pancreatic cancer. Genes Chromosomes Cancer 20, 383–391 (1997).
Fukushige, S. et al. Frequent gain of copy number on the long arm of chromosome 20 in human pancreatic adenocarcinoma. Genes Chromosomes Cancer 19, 161–169 (1997).
Curtis, L. J. et al. Amplification of DNA sequences from chromosome 19q31.1 in human pancreatic cell line. Genomics 53, 42–55 (1998).
Ghadimi, M. B. et al. Specific chromosomal aberrations and amplification of the AIB1 nuclear receptor coactivator gene in pancreatic carcinomas. Am. J. Pathol. 154, 525–536 (1999).
Schleger, C., Arens, N., Zentraf, H., Bleyl, U. & Verbeke, C. Identification of frequent chromosomal aberrations in ductal adenocarcinoma of the pancreas by comparative genomic hybridization (CGH). J. Pathol. 191, 27–32 (2000).
Shiraishi, K. et al. A comparison of DNA copy number changes detected by comparative genomic hybridization in malignancies of the liver, biliary tract and pancreas. Oncology 60, 151–161 (2001).
Harada, T. et al. Detection of genetic alterations in pancreatic cancers by comparative genomic hybridization coupled with tissue microdissection and degenerate oligonucleotide primed polymerase chain reaction. Oncology 62, 251–258 (2002).
Mahlamäki, E. H. et al. Frequent amplification of 8q24, 11q, 17q and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer 35, 353–358 (2002).
Lin, M., Cai, D. & Luo, M. Identification of chromosomal imbalances in pancreatic carcinoma using comparative genomic hybridization. Chin. Med. J. (Engl.) 116, 1156–1160 (2003).
Aguirre, A. J. et al. High resolution characterization of the pancreatic adenocarcinoma genome. Proc. Natl Acad. Sci. USA 101, 9067–9072 (2004).
Mahlamäki, E. H. et al. High resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia 6, 432–439 (2004).
Bashyam, M. D. et al. Array-based comparative genomic hybridization identifies localized DNA amplifications and homozygous deletions in pancreatic cancer. Neoplasia 7, 556–562 (2005).
Gysin, S., Rickert, P., Kastury, K. & McMahon, M. Analysis of genomic DNA alterations and mRNA expression patterns in a panel of human pancreatic cancer cell lines. Genes Chromosomes Cancer 44, 37–51 (2005).
Nowak, N. J. et al. Genome-wide aberrations in pancreatic adenocarcinoma. Cancer Genet. Cytogenet. 161, 36–50 (2005).
Harada, T., Chelala, C., Crnogorac-Jurcevic, T. & Lemoine, N. R. Genome-wide analysis of pancreatic cancer using microarray-based techniques. Pancreatology 9, 13–24 (2009).
Calhoun, E. S. et al. Identifying allelic loss and homozygous deletions in pancreatic cancer without matched normals using high-density single-nucleotide polymorphism arrays. Cancer Res. 66, 7920–7928 (2006).
Harada, T. et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene 27, 1951–1960 (2008).
Stoecklein, N. H. et al. Copy number of chromosome 17 but not HER2 amplification predicts clinical outcome of patients with pancreatic ductal adenocarincoma. J. Clin. Oncol. 22, 4737–4745 (2004).
Forbes, S. A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer Nucleic Acids Res. 39, D945–D950 (2011).
Acknowledgements
N. Samuel is supported by an award from the Frank Fletcher Memorial Fund. T. J. Hudson is the recipient of a Senior Investigator Award from the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Research and Innovation.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to all aspects of this Review.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Samuel, N., Hudson, T. The molecular and cellular heterogeneity of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 9, 77–87 (2012). https://doi.org/10.1038/nrgastro.2011.215
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2011.215
This article is cited by
-
Metabolic responses to the occurrence and chemotherapy of pancreatic cancer: biomarker identification and prognosis prediction
Scientific Reports (2024)
-
DDX18 drives tumor immune escape through transcription-activated STAT1 expression in pancreatic cancer
Oncogene (2023)
-
GNG7 and ADCY1 as diagnostic and prognostic biomarkers for pancreatic adenocarcinoma through bioinformatic-based analyses
Scientific Reports (2021)
-
Pancreatic cancer drug-sensitivity predicted by synergy of p53-Activator Wnt Inhibitor-2 (PAWI-2) and protein biomarker expression
Investigational New Drugs (2021)
-
Validation Strategy for Ultrasensitive Mutation Detection
Molecular Diagnosis & Therapy (2018)