Abstract
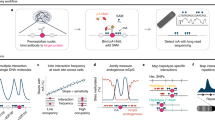
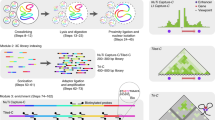
Chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) is a robust method for capturing genome-wide chromatin interactions. Unlike other 3C-based methods, it includes a chromatin immunoprecipitation (ChIP) step that enriches for interactions mediated by specific target proteins. This unique feature allows ChIA-PET to provide the functional specificity and higher resolution needed to detect chromatin interactions, which chromosome conformation capture (3C)/Hi-C approaches have not achieved. The original ChIA-PET protocol generates short paired-end tags (2 × 20 base pairs (bp)) to detect two genomic loci that are far apart on linear chromosomes but are in spatial proximity in the folded genome. We have improved the original approach by developing long-read ChIA-PET, in which the length of the paired-end tags is increased (up to 2 × 250 bp). The longer PET reads not only improve the tag-mapping efficiency but also increase the probability of covering phased single-nucleotide polymorphisms (SNPs), which allows haplotype-specific chromatin interactions to be identified. Here, we provide the detailed protocol for long-read ChIA-PET that includes cell fixation and lysis, chromatin fragmentation by sonication, ChIP, proximity ligation with a bridge linker, Tn5 tagmentation, PCR amplification and high-throughput sequencing. For a well-trained molecular biologist, it typically takes 6 d from cell harvesting to the completion of library construction, up to a further 36 h for DNA sequencing and <20 h for processing of raw sequencing reads.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Cremer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2, 292–301 (2001).
Bickmore, W.A. The spatial organization of the human genome. Annu. Rev. Genomics Hum. Genet. 14, 67–84 (2013).
Cullen, K.E., Kladde, M.P. & Seyfred, M.A. Interaction between transcription regulatory regions of prolactin chromatin. Science 261, 203–206 (1993).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Dostie, J. & Dekker, J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat. Protoc. 2, 988–1002 (2007).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Fullwood, M.J. et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 (2009).
Sati, S. & Cavalli, G. Chromosome conformation capture technologies and their impact in understanding genome function. Chromosoma (2016).
Li, G. et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 (2012).
Zhang, Y. et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature 504, 306–310 (2013).
Kieffer-Kwon, K.R. et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155, 1507–1520 (2013).
Handoko, L. et al. CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 (2011).
Tang, Z. et al. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell 163, 1611–1627 (2015).
DeMare, L.E. et al. The genomic landscape of cohesin-associated chromatin interactions. Genome Res. 23, 1224–1234 (2013).
Ji, X. et al. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18, 262–275 (2015).
Chepelev, I., Wei, G., Wangsa, D., Tang, Q. & Zhao, K. Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 (2012).
Heidari, N. et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 24, 1905–1917 (2014).
Leung, D. et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature 518, 350–354 (2015).
McDaniell, R. et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science 328, 235–239 (2010).
Goh, Y. et al. Chromatin interaction analysis with paired-end Tag sequencing (ChIA-PET) for mapping chromatin interactions and understanding transcription regulation. J. Vis. Exp. 62, 3770 (2012).
Jackson, V. Studies on histone organization in the nucleosome using formaldehyde as a reversible cross-linking agent. Cell 15, 945–954 (1978).
Dekker, J., Marti-Renom, M.A. & Mirny, L.A. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 14, 390–403 (2013).
Jin, F. et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013).
Rao, S.S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Orlando, V., Strutt, H. & Paro, R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11, 205–214 (1997).
Zeng, P.Y., Vakoc, C.R., Chen, Z.C., Blobel, G.A. & Berger, S.L. In vivo dual cross-linking for identification of indirect DNA-associated proteins by chromatin immunoprecipitation. Biotechniques 41 694, 696, 698 (2006).
Zhang, J. et al. ChIA-PET analysis of transcriptional chromatin interactions. Methods 58, 289–299 (2012).
Toews, J., Rogalski, J.C., Clark, T.J. & Kast, J. Mass spectrometric identification of formaldehyde-induced peptide modifications under in vivo protein cross-linking conditions. Anal. Chim. Acta 618, 168–183 (2008).
Fullwood, M.J. & Ruan, Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell Biochem. 107, 30–39 (2009).
Li, G. et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 11, R22 (2010).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A.R. & Hall, I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
Y.R. is supported by the Director Innovation Fund of The Jackson Laboratory; grant nos. NCI R01 CA186714, NHGRI R25HG007631, and NIDDK U54DK107967 (4DN); and the Roux family endowment. X.L. is supported in part by China's '111 project' (grant no. B07041). G.L. is supported by the National Natural Science Foundation of China (grant no. 91440114) and the Fundamental Research Funds for the Central Universities (grant no. 2662014PY001).
Author information
Authors and Affiliations
Contributions
X.L. and Y.R. designed the experimental part of the protocol; O.J.L. and G.L. designed the data-processing pipeline and analyzed the data with assistance from S.Z.T. and Z.T.; X.L. implemented the protocol with assistance from P.W., M.Z., D.W., E.P. and J.J.Z.; X.L., O.J.L. and Y.R. wrote the manuscript with input from G.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Software 1
Perl script 1: prepare_reads_single.pl. (TXT 5 kb)
Supplementary Software 2
Perl script 2: prepare_reads_pair.pl. (TXT 1 kb)
Supplementary Software 3
Perl script 3: clustering_bridge.pl. (TXT 2 kb)
Supplementary Software 4
Python script 1: fetch_base_snp_v081.py. (TXT 1 kb)
Rights and permissions
About this article
Cite this article
Li, X., Luo, O., Wang, P. et al. Long-read ChIA-PET for base-pair-resolution mapping of haplotype-specific chromatin interactions. Nat Protoc 12, 899–915 (2017). https://doi.org/10.1038/nprot.2017.012
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.012
This article is cited by
-
ChIPr: accurate prediction of cohesin-mediated 3D genome organization from 2D chromatin features
Genome Biology (2024)
-
Comparative study on chromatin loop callers using Hi-C data reveals their effectiveness
BMC Bioinformatics (2024)
-
Hi-Tag: a simple and efficient method for identifying protein-mediated long-range chromatin interactions with low cell numbers
Science China Life Sciences (2024)
-
3D organization of regulatory elements for transcriptional regulation in Arabidopsis
Genome Biology (2023)
-
3D genomics and its applications in precision medicine
Cellular & Molecular Biology Letters (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.