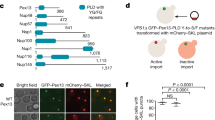
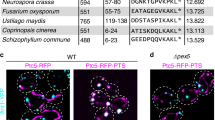
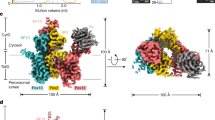
Abstract
Here we describe a protocol to dissect the peroxisomal matrix protein import pathway using a cell-free in vitro system. The system relies on a postnuclear supernatant (PNS), which is prepared from rat/mouse liver, to act as a source of peroxisomes and cytosolic components. A typical in vitro assay comprises the following steps: (i) incubation of the PNS with an in vitro–synthesized 35S-labeled reporter protein; (ii) treatment of the organelle suspension with a protease that degrades reporter proteins that have not associated with peroxisomes; and (iii) SDS–PAGE/autoradiography analysis. To study transport of proteins into peroxisomes, it is possible to use organelle-resident proteins that contain a peroxisomal targeting signal (PTS) as reporters in the assay. In addition, a receptor (PEX5L/S or PEX5L.PEX7) can be used to report the dynamics of shuttling proteins that mediate the import process. Thus, different but complementary perspectives on the mechanism of this pathway can be obtained. We also describe strategies to fortify the system with recombinant proteins to increase import yields and block specific parts of the machinery at a number of steps. The system recapitulates all the steps of the pathway, including mono-ubiquitination of PEX5L/S at the peroxisome membrane and its ATP-dependent export back into the cytosol by PEX1/PEX6. An in vitro import(/export) experiment can be completed in 24 h.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Vögtle, F.-N. et al. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139, 428–439 (2009).
Karamyshev, A.L. et al. Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell 156, 146–157 (2014).
Rodrigues, T.A. et al. A PEX7-centered perspective on the peroxisomal targeting signal type 2-mediated protein import pathway. Mol. Cell. Biol. 34, 2917–2928 (2014).
Paila, Y.D. et al. Multi-functional roles for the polypeptide transport associated domains of Toc75 in chloroplast protein import. Elife 5 (2016).
Miura, S., Miyazawa, S., Osumi, T., Hashimoto, T. & Fujiki, Y. Post-translational import of 3-ketoacyl-CoA thiolase into rat liver peroxisomes. J. Biochem. 115, 1064–1068 (1994).
Small, G.M., Imanaka, T., Shio, H. & Lazarow, P.B. Efficient association of in vitro translation products with purified stable Candida tropicalis peroxisomes. Mol. Cell. Biol. 7, 1848–1855 (1987).
Thieringer, R., Shio, H., Han, Y.S., Cohen, G. & Lazarow, P.B. Peroxisomes in Saccharomyces cerevisiae: immunofluorescence analysis and import of catalase A into isolated peroxisomes. Mol. Cell. Biol. 11, 510–522 (1991).
Brickner, D.G., Harada, J.J. & Olsen, L.J. Protein transport into higher plant peroxisomes. In vitro import assay provides evidence for receptor involvement. Plant Physiol. 113, 1213–1221 (1997).
Brickner, D.G. & Olsen, L.J. Nucleotide triphosphates are required for the transport of glycolate oxidase into peroxisomes. Plant Physiol. 116, 309–317 (1998).
Grou, C.P. et al. The peroxisomal protein import machinery - a case report of transient ubiquitination with a new flavor. Cell. Mol. Life Sci. 66, 254–262 (2009).
Kim, P.K. & Hettema, E.H. Multiple pathways for protein transport to peroxisomes. J. Mol. Biol. 427, 1176–1190 (2015).
Baker, A. & Paudyal, R. The life of the peroxisome: from birth to death. Curr. Opin. Plant Biol. 22, 39–47 (2014).
Rodrigues, T.A., Grou, C.P. & Azevedo, J.E. Revisiting the intraperoxisomal pathway of mammalian PEX7. Sci. Rep. 5, 11806 (2015).
Freitas, M.O. et al. The peroxisomal protein import machinery displays a preference for monomeric substrates. Open Biol. 5, 140236 (2015).
Alexson, S.E.H., Fujiki, Y., Shio, H. & Lazarow, P.B. Partial disassembly of peroxisomes. J. Cell Biol. 101, 294–305 (1985).
Brocard, C. & Hartig, A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim. Biophys. Acta 1763, 1565–1573 (2006).
Oliveira, M.E., Gouveia, A.M., Pinto, R.A., Sá-Miranda, C. & Azevedo, J.E. The energetics of Pex5p-mediated peroxisomal protein import. J. Biol. Chem. 278, 39483–39488 (2003).
Gouveia, A.M. et al. Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J. Biol. Chem. 278, 226–232 (2003).
Gouveia, A.M., Guimarães, C.P., Oliveira, M.E., Sá-Miranda, C. & Azevedo, J.E. Insertion of Pex5p into the peroxisomal membrane is cargo protein-dependent. J. Biol. Chem. 278, 4389–4392 (2003).
Gouveia, A.M., Reguenga, C., Oliveira, M.E., Sa-Miranda, C. & Azevedo, J.E. Characterization of peroxisomal Pex5p from rat liver. Pex5p in the Pex5p-Pex14p membrane complex is a transmembrane protein. J. Biol. Chem. 275, 32444–32451 (2000).
Carvalho, A.F. et al. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 (2007).
Miyata, N. & Fujiki, Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 (2005).
Grou, C.P. et al. Properties of the ubiquitin-pex5p thiol ester conjugate. J. Biol. Chem. 284, 10504–10513 (2009).
Bhogal, M.S., Lanyon-Hogg, T., Johnston, K.A., Warriner, S.L. & Baker, A. Covalent label transfer between peroxisomal importomer components reveals export-driven import interactions. J. Biol. Chem. 291, 2460–2468 (2016).
Platta, H.W., Grunau, S., Rosenkranz, K., Girzalsky, W. & Erdmann, R. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 (2005).
Alencastre, I.S. et al. Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway. J. Biol. Chem. 284, 27243–27251 (2009).
Francisco, T. et al. A cargo-centered perspective on the PEX5-mediated peroxisomal protein import pathway. J. Biol. Chem. 288, 29151–29159 (2013).
Williams, C.P. et al. The Peroxisomal Targeting Signal 1 in sterol carrier protein 2 is autonomous and essential for receptor recognition. BMC Biochem. 12, 12 (2011).
Williams, C., van den Berg, M., Sprenger, R.R. & Distel, B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 (2007).
Grou, C.P. et al. Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on the ubiquitin-peroxin 5 (PEX5) thioester conjugate. J. Biol. Chem. 287, 12815–12827 (2012).
Debelyy, M.O. et al. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 286, 28223–28234 (2011).
Leighton, F., Poole, B., Lazarow, P.B. & De Duve, C. The synthesis and turnover of rat liver peroxisomes. I. Fractionation of peroxisome proteins. J. Cell Biol. 41, 521–535 (1969).
Grou, C.P. et al. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J. Biol. Chem. 283, 14190–14197 (2008).
Okumoto, K. et al. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 12, 1067–1083 (2011).
Wendland, M. & Subramani, S. Cytosol-dependent peroxisomal protein import in a permeabilized cell system. J. Cell Biol. 120, 675–685 (1993).
Koepke, J.I. et al. Restoration of peroxisomal catalase import in a model of human cellular aging. Traffic 8, 1590–1600 (2007).
Terlecky, S.R., Legakis, J.E., Hueni, S.E. & Subramani, S. Quantitative analysis of peroxisomal protein import. Exp. Cell Res. 263, 98–106 (2001).
Pinto, M.P. et al. The import competence of a peroxisomal membrane protein is determined by Pex19p before the docking step. J. Biol. Chem. 281, 34492–34502 (2006).
Ghosh, D. & Berg, J.M. A proteome-wide perspective on peroxisome targeting signal 1(PTS1)-Pex5p affinities. J. Am. Chem. Soc. 132, 3973–3979 (2010).
Miyata, N., Okumoto, K., Mukai, S., Noguchi, M. & Fujiki, Y. AWP1/ZFAND6 functions in Pex5 export by interacting with cys-monoubiquitinated Pex5 and Pex6 AAA ATPase. Traffic 13, 168–183 (2012).
Croes, K., Foulon, V., Casteels, M., Van Veldhoven, P.P. & Mannaerts, G.P. Phytanoyl-CoA hydroxylase: recognition of 3-methyl-branched acyl-coAs and requirement for GTP or ATP and Mg(2+) in addition to its known hydroxylation cofactors. J. Lipid Res. 41, 629–636 (2000).
Miura, S. et al. Biosynthesis and intracellular transport of enzymes of peroxisomal beta-oxidation. J. Biol. Chem. 259, 6397–6402 (1984).
Schram, A.W. et al. Biosynthesis and maturation of peroxisomal beta-oxidation enzymes in fibroblasts in relation to the Zellweger syndrome and infantile Refsum disease. Proc. Natl. Acad. Sci. USA 83, 6156–6158 (1986).
Ossendorp, B.C. et al. Tissue-specific distribution of a peroxisomal 46-kDa protein related to the 58-kDa protein (sterol carrier protein x; sterol carrier protein 2/3-oxoacyl-CoA thiolase). Arch. Biochem. Biophys. 334, 251–260 (1996).
Braverman, N., Dodt, G., Gould, S.J. & Valle, D. An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Hum. Mol. Genet. 7, 1195–1205 (1998).
Lazarow, P.B. The import receptor Pex7p and the PTS2 targeting sequence. Biochim. Biophys. Acta 1763, 1599–1604 (2006).
Kunze, M. et al. Structural requirements for interaction of peroxisomal targeting signal 2 and its receptor PEX7. J. Biol. Chem. 286, 45048–45062 (2011).
Otera, H. et al. Peroxisome targeting signal type 1 (PTS1) receptor is involved in import of both PTS1 and PTS2: studies with PEX5-defective CHO cell mutants. Mol. Cell. Biol. 18, 388–399 (1998).
Fujiki, Y., Matsuzono, Y., Matsuzaki, T. & Fransen, M. Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim. Biophys. Acta 1763, 1639–1646 (2006).
Freitas, M.O. et al. PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J. Biol. Chem. 286, 40509–40519 (2011).
Dodt, G. et al. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat. Genet. 9, 115–125 (1995).
Gatto, G. & Geisbrecht, B. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat. Struct. Biol. 7, 1091–1095 (2000).
Szilard, R.K. & Rachubinski, R.A. Tetratricopeptide repeat domain of Yarrowia lipolytica Pex5p is essential for recognition of the type 1 peroxisomal targeting signal but does not confer full biological activity on Pex5p. Biochem. J. 346, 177–184 (2000).
Carvalho, A.F. et al. Functional characterization of two missense mutations in Pex5p - C11S and N526K. Biochim. Biophys. Acta 1773, 1141–1148 (2007).
Nordgren, M. et al. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy 11, 1326–1340 (2015).
Carvalho, A.F. et al. The N-terminal half of the peroxisomal cycling receptor Pex5p is a natively unfolded domain. J. Mol. Biol. 356, 864–875 (2006).
Schliebs, W. et al. Recombinant human peroxisomal targeting signal receptor PEX5: structural basis for interaction of PEX5 with PEX14. J. Biol. Chem. 274, 5666–5673 (1999).
Otera, H. et al. Peroxisomal targeting signal receptor Pex5p interacts with cargoes and import machinery components in a spatiotemporally differentiated manner: conserved Pex5p WXXXF/Y motifs are critical for matrix protein import. Mol. Cell. Biol. 22, 1639–1655 (2002).
Costa-Rodrigues, J. et al. The N terminus of the peroxisomal cycling receptor, Pex5p, is required for redirecting the peroxisome-associated peroxin back to the cytosol. J. Biol. Chem. 279, 46573–46579 (2004).
Hwang, S.T. & Schatz, G. Translocation of proteins across the mitochondrial inner membrane, but not into the outer membrane, requires nucleoside triphosphates in the matrix. Proc. Natl. Acad. Sci. USA 86, 8432–8436 (1989).
Haas, A.L., Warms, J. & Rose, I.A. Ubiquitin adenylate: structure and role in ubiquitin activation. Biochemistry 22, 4388–4394 (1983).
Pickart, C.M. & Rose, I.A. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J. Biol. Chem. 260, 7903–7910 (1985).
Costa-Rodrigues, J. et al. Pex5p, the peroxisomal cycling receptor, is a monomeric non-globular protein. J. Biol. Chem. 280, 24404–24411 (2005).
Ferro, A. et al. NEDD8: a new ataxin-3 interactor. Biochim. Biophys. Acta 1773, 1619–1627 (2007).
Fransen, M., Wylin, T., Brees, C., Mannaerts, G.P. & Van Veldhoven, P.P. Human pex19p binds peroxisomal integral membrane proteins at regions distinct from their sorting sequences. Mol. Cell. Biol. 21, 4413–4424 (2001).
Acknowledgements
We thank M. Fransen, Katholieke Universiteit-Leuven, for critical comments on the manuscript and for the plasmid encoding histidine-tagged PEX19. We thank P. van Veldhoven, Katholieke Universiteit-Leuven, and P. Maciel, Universidade do Minho, for the expression plasmids encoding prePHYH and GST-Ub, respectively. This work was funded by FEDER—Fundo Europeu de Desenvolvimento Regional through the COMPETE 2020—Operacional Programme for Competitiveness and Internationalization (POCI), Portugal 2020, Portugal's FCT—Fundação para a Ciência e a Tecnologia/ Ministério da Ciência, Tecnologia e Inovação in the framework of the projects 'The molecular mechanism of protein import into peroxisomes' (FCOMP-01-0124-FEDER-019731-PTDC/BIA-BCM/118577/2010), 'Institute for Research and Innovation in Health Sciences' (POCI-01-0145-FEDER-007274) and 'The molecular mechanisms of peroxisome biogenesis' (PTDC /BEX-BC M/2311/2014) and Norte 2020—Programa Operacional Regional do Norte, under the application of the 'Porto Neurosciences and Neurologic Disease Research Initiative at i3S (NORTE-01-0145-FEDER-000008)', awarded to J.E.A. T.A.R., T.F., A.F.D. and C.P.G. were supported by Fundação para a Ciência e a Tecnologia, Programa Operacional Potencial Humano do QREN and Fundo Social Europeu.
Author information
Authors and Affiliations
Contributions
T.A.R., T.F., C.P.G. and J.E.A. designed research. T.A.R., T.F., A.F.D. and A.G.P. performed the experiments. T.A.R., T.F., A.F.D., A.G.P., C.P.G. and J.E.A. wrote and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Rodrigues, T., Francisco, T., Dias, A. et al. A cell-free organelle-based in vitro system for studying the peroxisomal protein import machinery. Nat Protoc 11, 2454–2469 (2016). https://doi.org/10.1038/nprot.2016.147
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.147
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.