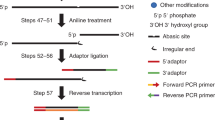
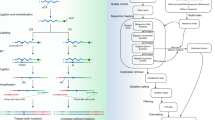
Abstract
Cytosine methylation within RNA is common, but its full scope and functions are poorly understood, as the RNA targets of most mammalian cytosine RNA methyltransferases (m5C-RMTs) remain uncharacterized. To enable their characterization, we developed a mechanism-based method for transcriptome-wide m5C-RMT target profiling. All characterized mammalian m5C-RMTs form a reversible covalent intermediate with their cytosine substrate—a covalent linkage that is trapped when conducted on the cytosine analog 5-azacytidine (5-aza-C). We used this property to develop Aza-immunoprecipitation (Aza-IP), a methodology to form stable m5C-RMT-RNA linkages in cell culture, followed by IP and high-throughput sequencing, to identify direct RNA substrates of m5C-RMTs. Remarkably, a cytosine-to-guanine (C→G) transversion occurs specifically at target cytosines, allowing the simultaneous identification of the precise target cytosine within each RNA. Thus, Aza-IP reports only direct RNA substrates and the C→G transversion provides an important criterion for target cytosine identification, which is not available in alternative approaches. Here we present a step-by-step protocol for Aza-IP and downstream analysis, designed to reveal identification of substrate RNAs and precise cytosine targets of m5C-RMTs. The entire protocol takes 40–50 d to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Machnicka, M.A. et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res. 41, D262–D267 (2013).
Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 34, 721–733 (2006).
Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874 (2011).
Mercer, T.R. & Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 20, 300–307 (2013).
Kellner, S., Burhenne, J. & Helm, M. Detection of RNA modifications. RNA Biol. 7, 237–247 (2010).
Motorin, Y., Muller, S., Behm-Ansmant, I. & Branlant, C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 425, 21–53 (2007).
Affymetrix ENCODE Transcriptome Project; Cold Spring Harbor Laboratory ENCODE Transcriptome Project. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature 457, 1028–1032 (2009).
Dominissini, D. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206 (2012).
Meyer, K.D. et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646 (2012).
Edelheit, S., Schwartz, S., Mumbach, M.R., Wurtzel, O. & Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 9, e1003602 (2013).
Gowda, M. et al. Genome-wide characterization of methylguanosine-capped and polyadenylated small RNAs in the rice blast fungus Magnaporthe oryzae. Nucleic Acids Res. 38, 7558–7569 (2010).
Khoddami, V. & Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 31, 458–464 (2013).
Motorin, Y., Lyko, F. & Helm, M. 5-methylcytosine in RNA: detection, enzymatic formation and biological functions. Nucleic Acids Res. 38, 1415–1430 (2010).
Santi, D.V., Garrett, C.E. & Barr, P.J. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell 33, 9–10 (1983).
Lu, L.W., Chiang, G.H., Medina, D. & Randerath, K. Drug effects on nucleic acid modification. I. A specific effect of 5-azacytidine on mammalian transfer RNA methylation in vivo. Biochem. Biophys. Res. Commun. 68, 1094–1101 (1976).
Lu, L.J. & Randerath, K. Effects of 5-azacytidine on transfer RNA methyltransferases. Cancer Res. 39, 940–949 (1979).
Schaefer, M., Hagemann, S., Hanna, K. & Lyko, F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res. 69, 8127–8132 (2009).
Oki, Y., Aoki, E. & Issa, J.P. Decitabine–bedside to bench. Crit. Rev. Oncol. Hematol. 61, 140–152 (2007).
Squires, J.E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 40, 5023–5033 (2012).
Hussain, S. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 4, 255–261 (2013).
Tuorto, F. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 19, 900–905 (2012).
Abbasi-Moheb, L. et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 847–855 (2012).
Khan, M.A. et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 90, 856–863 (2012).
Martinez, F.J. et al. Whole-exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 49, 380–385 (2012).
Frye, M. & Watt, F.M. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 16, 971–981 (2006).
Blanco, S. et al. The RNA-methyltransferase Misu (NSun2) poises epidermal stem cells to differentiate. PLoS Genet. 7, e1002403 (2011).
Chan, C.T. et al. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 6, e1001247 (2010).
Becker, M. et al. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res. 40, 11648–11658 (2012).
Chan, C.T. et al. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 3, 937 (2012).
Rai, K. et al. Dnmt2 functions in the cytoplasm to promote liver, brain, and retina development in zebrafish. Genes Dev. 21, 261–266 (2007).
Fonagy, A. et al. Cell cycle regulated expression of nucleolar antigen P120 in normal and transformed human fibroblasts. J. Cell Physiol. 154, 16–27 (1993).
Frye, M. et al. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 289, 71–80 (2010).
Okamoto, M. et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 31, 660–671 (2012).
Job, B. et al. Genomic aberrations in lung adenocarcinoma in never smokers. PLoS ONE 5, e15145 (2010).
Doll, A. & Grzeschik, K.H. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet Cell Genet. 95, 20–27 (2001).
Harris, T., Marquez, B., Suarez, S. & Schimenti, J. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol. Reprod. 77, 376–382 (2007).
Jurkowski, T.P. et al. Human DNMT2 methylates tRNAAsp molecules using a DNA methyltransferase-like catalytic mechanism. RNA 14, 1663–1670 (2008).
King, M.Y. & Redman, K.L. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry 41, 11218–11225 (2002).
Sharma, S., Yang, J., Watzinger, P., Kotter, P. & Entian, K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 41, 9062–9076 (2013).
Hussain, S. et al. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 186, 27–40 (2009).
Chi, L. & Delgado-Olguin, P. Expression of NOL1/NOP2/sun domain (Nsun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr. Patterns 13, 319–327 (2013).
Nix, D.A., Courdy, S.J. & Boucher, K.M. Empirical methods for controlling false positives and estimating confidence in ChIP-seq peaks. BMC Bioinformatics 9, 523 (2008).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Koboldt, D.C. et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25, 2283–2285 (2009).
Robinson, J.T. et al. Integrative Genomics Viewer. Nat. Biotechnol. 29, 24–26 (2011).
Klages, N., Zufferey, R. & Trono, D. A stable system for the high-titer production of multiply attenuated lentiviral vectors. Mol. Ther. 2, 170–176 (2000).
Acknowledgements
We thank A. Schlichter, T. Parnell, A. Yerra, B. Dalley, N. Moss, K. Basham, D. Nix, T. Mosbruger and B. Milash for their helpful comments and suggestions for improving the manuscript. This work was supported by HHMI and the Samuel Waxman Cancer Research Foundation, as well as by the National Cancer Institute CA24014 (for core facilities). We thank V. Planelles (University of Utah) for pPR-lentiviral expression plasmid.
Author information
Authors and Affiliations
Contributions
B.R.C. and V.K. together conceived Aza-IP and the general experimental design. V.K. was involved in detailed design, implementation of all experiments and bioinformatic analysis for Aza-IP and RNA bisulfite sequencing. V.K. and B.R.C. wrote the main text. V.K. wrote the remaining text. V.K. designed and made all figures, with comments from B.R.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Schematic of the 2-step PCR amplification of NSUN2 mRNA.
First, cDNA is made using the oligo-dT primer (Step 5). Next, the F1 and R primers are used to produce amplicon 1 (Steps 6-8), which is used as template for the F2 and R primers to produce amplicon 2 (Steps 9-11). Amplicon 2 contains all the elements of the designed construct (Supplementary Data). Sequences of the F1, F2 and R primers are provided in the MATERIALS section.
Supplementary information
Supplementary Figure 1
Schematic of the 2-step PCR amplification of NSUN2 mRNA. (PDF 137 kb)
Supplementary Data
Supplementary Table 1: CDS and protein sequences of the NSUN2 variant 1. Supplementary Table 2: Designed NSUN2 amplicon for restriction enzyme digestion and cloning into lentiviral vector. (PDF 178 kb)
Rights and permissions
About this article
Cite this article
Khoddami, V., Cairns, B. Transcriptome-wide target profiling of RNA cytosine methyltransferases using the mechanism-based enrichment procedure Aza-IP. Nat Protoc 9, 337–361 (2014). https://doi.org/10.1038/nprot.2014.014
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.014
This article is cited by
-
Preclinical efficacy of azacitidine and venetoclax for infant KMT2A-rearranged acute lymphoblastic leukemia reveals a new therapeutic strategy
Leukemia (2023)
-
Advances in RNA cytosine-5 methylation: detection, regulatory mechanisms, biological functions and links to cancer
Biomarker Research (2020)
-
RNA-modifying enzymes and their function in a chromatin context
Nature Structural & Molecular Biology (2019)
-
Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain
Genome Biology (2017)
-
Detecting RNA modifications in the epitranscriptome: predict and validate
Nature Reviews Genetics (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.