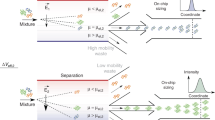
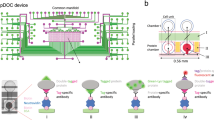
Abstract
This protocol describes regional photopatterning of polyacrylamide gels in glass microfluidic devices as a platform for seamless integration of multiple assay steps. The technology enables rapid, automated protein immunoblotting, demonstrated in this study for native western blotting. The fabrication procedure is straightforward and requires approximately 3 h from the start of gel photopatterning to completion of native protein western blotting, a substantial time savings over slab-gel immunoblotting. The assay itself requires less than 5 min. Importantly, all assay stages are programmably controlled by a high-voltage power supply and monitored by an epifluorescence microscope equipped with a charge-coupled device camera. Our approach overcomes severe limitations associated with conventional immunoblotting, including multiple steps requiring manual intervention, low throughput and substantial consumption of reagents. We also describe a simple chemical recycling protocol so that glass chips can be reused. The fabrication technique described forms the basis for a diverse suite of bioanalytical tools, including DNA/RNA blotting and multidimensional separations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Whitesides, G.M. The origins and the future of microfluidics. Nature 442, 368–373 (2006).
El-Ali, J.E., Sorger, P.K. & Jensen, K.F. Cells on chips. Nature 442, 403–411 (2006).
Craighead, H. Future lab-on-a-chip technologies for integrating individual molecules. Nature 442, 387–393 (2006).
Janasek, D., Franzke, J. & Manz, A. Scaling and the design of miniaturized chemical-analysis systems. Nature 442, 374–380 (2006).
Eijkel, J.C.T. & van den Berg, A. Nanofluidics: what is it and what can we expect from it? Microfluid. Nanofluid. 1, 249–267 (2005).
Yager, P. et al. Microfluidic diagnostic technologies for global public health. Nature 442, 412–418 (2006).
Thorsen, T., Maerkl, S.J. & Quake, S.R. Microfluidic large-scale integration. Science 298, 580–584 (2002).
Huang, B. et al. Counting low-copy number proteins in a single cell. Science 315, 81–84 (2007).
Liu, J., Yang, S., Lee, C.S. & DeVoe, D. Polyacrylamide gel plugs enabling 2-D microfluidic protein separations via isoelectric focusing and multiplexed sodium dodecyl sulfate gel electrophoresis. Electrophoresis 29, 2241–2250 (2008).
Das, C., Zhang, J., Denslow, N.D. & Fan, Z.H. Integration of isoelectric focusing with multi-channel gel electrophoresis by using microfluidic pseudo-valves. Lab. Chip. 7, 1806–1812 (2007).
Song, S., Singh, A.K., & Kirby, B.J. Electrophoretic concentration of proteins at laser-patterned nanoporous membranes in microchips. Anal. Chem. 76, 4589–4592 (2004).
Hatch, A.V., Herr, A.E., Throckmorton, D.J., Brennan, J.S. & Singh, A.K. Integrated preconcentration SDS-PAGE of proteins in microchips using photopatterned cross-linked polyacrylamide gels. Anal. Chem. 78, 4976–4984 (2006).
Herr, A.E. et al. Microfluidic immunoassays as rapid saliva-based clinical diagnostics. Proc. Natl Acad. Sci. USA 104, 5268–5273 (2007).
Peterson, D.S. Solid supports for micro analytical systems. Lab. Chip. 5, 132–139 (2005).
Svec, F. My favorite materials: porous polymer monoliths. J. Sep. Sci. 32, 3–9 (2009).
Schägger, H. Tricine-SDS-PAGE. Nat. Protoc. 1, 16–22 (2006).
Wittig, I., Braun, H.P. & Schägger, H. Blue native PAGE. Nat. Protoc. 1, 418–428 (2006).
Kinoshita, E., Kinoshita-Kikuta, E. & Koike, T. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4, 1513–1521 (2009).
Stead, J.A. & McDowall, K.J. Two-dimensional gel electrophoresis for identifying proteins that bind DNA or RNA. Nat. Protoc. 2, 1839–1848 (2007).
He, M. & Herr, A.E. Microfluidic polyacrylamide gel electrophoresis with in-situ immunoblotting for native protein analysis. Anal. Chem. 81, 8177–8184 (2009).
Fan, A.C. et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat. Med. 15, 566–571 (2009).
He, M. & Herr, A.E. Polyacrylamide gel photopatterning enables automated protein immunoblotting in a two-dimensional microdevice. J. Am. Chem. Soc. 132, 2512–2513 (2010).
Kurien, B.T. & Scofield, R.H. Protein Blotting and Detection: Methods and Protocols 1–33 (Humana Press, New York, USA, 2009).
Yamada, M., Mao, P., Fu, J. & Han, J. Rapid quantification of disease-marker proteins using continuous-flow immunoseparation in a nanosieve fluidic device. Anal. Chem. 81, 7067–7074 (2009).
Fu, J., Mao, P. & Han, J. Artificial molecular sieves and filters: a new paradigm for biomolecule separation. Trends Biotech. 26, 311–320 (2008).
Han, J. & Craighead, H.G. Separation of long DNA molecules in a microfabricated entropic trap array. Science 288, 1026–1029 (2000).
Fu, J. et al. A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins. Nat. Nanotech. 2, 121–128 (2007).
Huang, L.R. et al. A DNA prism for high-speed continuous fractionation of large NDA molecules. Nat. Biotech. 20, 1048–1051 (2002).
Wirth, M.J. Separation media for microchips. Anal. Chem. 79, 800–808 (2007).
Zeng, Y., He, M. & Harrison, D.J. Microfluidic self-patterning of large-scale crystalline nanoarrays for high-throughput continuous DNA fractionation. Angew. Chem. Int. Ed. 47, 6388–6391 (2008).
Fu, J., Mao, P. & Han, J. Continuous-flow bioseparation using microfabricated anisotropic nanofluidic sieving structures. Nat. Protoc. 4, 1681–1698 (2009).
Xiong, L., Zhang, R. & Regnier, F.E. Potential of silica monolithic columns in peptide separations. J. Chromatogr. A 1030, 187–194 (2004).
Awada, C., Sato, T. & Takao, T. Affinity-trap polyacrylamide gel electrophoresis: a novel method of capturing specific proteins by electro-transfer. Anal. Chem. 82, 755–761 (2010).
Herr, A.E., Throckmorton, D.J., Davenport, A.A. & Singh, A.K. On-chip native gel electrophoresis-based immunoassays for tetanus antibody and toxin. Anal. Chem. 77, 585–590 (2005).
Herr, A.E. & Singh, A.K. Photopolymerized cross-linked polyacrylamide gels for on-chip protein sizing. Anal. Chem. 76, 4727–4733 (2004).
Hou, C. & Herr, A.E. Ultra-short separation length homogeneous electrophoretic immunoassays using on-chip discontinuous polyacrylamide gels. Anal. Chem. 82, 3343–3351 (2010).
Jemere, A.B., Oleschuk, R.D. & Harrison, D.J. Microchip-based capillary electrochromatography using packed beds. Electrophoresis 24, 3018–3025 (2003).
Oleschuk, R.D., Shultz-Lockyear, L.L., Ning, Y. & Harrison, D.J. Trapping of bead based reagents within microfluidic systems: on-chip solid-phase extraction and electrochromatography. Anal. Chem. 72, 585–590 (2000).
Yu, C., Xu, M., Svec, F. & Fréchet, J.M.J. Preparation of monolithic polymers with controlled porous properties for microfluidic chip applications using photoinitiated free-radical polymerization. J. Polym. Sci. Part A Polym. Chem. 40, 755–769 (2002).
Le Gac, S.L., Carlier, J., Camart, J.C., Olivé, C.C. & Rolando, C. Monoliths for microfluidic devices in proteomics. J. Chromatogr. B 808, 3–14 (2004).
Song, S., Singh, A.K., Shepodd, T.J. & Kirby, B.J. Microchip dialysis of proteins using in situ photopatterned nanoporous polymer membranes. Anal. Chem. 76, 2367–2373 (2004).
Towbin, H., Staehelin, T. & Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA 76, 4350–4354 (1979).
Wu, Y., Li, Q. & Chen, X.Z. Detecting protein-protein interactions by Far western blotting. Nat. Protoc. 2, 3278–3284 (2007).
Ciaccio, M.F., Wagner, J.P., Chuu, C.P., Lauffenburger, D.A., & Jones, R.B. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat. Methods 7, 148–155 (2010).
Kolli, M., Hamidouche, M., Bouaouadja, N. & Fantozzi, G., et al. HF etching effect on sandblasted soda-lime glass properties. J. Eur. Ceram. Soc. 29, 2697–2704 (2009).
He, Q. et al. Fabrication of 1D nanofluidic channels on glass substrate by wet etching and room-temperature bonding. Anal. Chim. Acta. 628, 1–8 (2008).
Iliescu, C., Chen, B. & Miao, J. On the wet etching of Pyrex glass. Sens. Actuators A 143, 154–161 (2008).
Lerch, M.A. & Jacobson, S.C. Electrokinetic fluid control in two-dimensional planar microfluidic devices. Anal. Chem. 79, 7485–7491 (2007).
Huang, L.R. et al. Generation of large-area tunable uniform electric fields in microfluidics arrays for rapid DNA separation. Tech. Dig. Int. El. Devices Meet. 1, 363–366 (2001).
Das, C., Fredrickson, C.K., Xia, Z. & Fan, Z.H. Device fabrication and integration with photodefinable microvalves for protein separation. Sensor Actuat. A Phys. 134, 271–277 (2007).
Sensabaugh, G.F. & Blake, E.T. Seminal plasma protein p30: simplified purification and evidence for identity with prostate-specific antigen. J. Urol. 144, 1523–1526 (1990).
Collins, T.J. ImageJ for microscopy. Biotechniques 43, S25–S30 (2007).
Acknowledgements
We acknowledge financial support from the University of California, Berkeley and the QB3/Rogers Family Foundation Award. Facilities and equipment support from UC Berkeley's Biomolecular Nanofabrication Center is also appreciated. PSA samples were a generous gift from D. Peehl, Stanford University. A.E.H. is an Alfred P. Sloan Foundation Research Fellow in chemistry.
Author information
Authors and Affiliations
Contributions
M.H. designed chips and conducted experiments. M.H. and A.E.H. analyzed data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Microscale immunoblot chip layout and dimensions (unit: mm). Channel depth is 20µm. The closed squares and circles designate areas for etching. The regions within the green and blue boxes are depicted further in Supplementary Figure 2 and Supplementary Figure 3, respectively. (PDF 217 kb)
Supplementary Fig. 2
Green box detail from Supplementary Figure 1 (unit: mm). Channel depth is 20µm. The closed square and triangles designate areas for etching. (PDF 211 kb)
Supplementary Fig. 3
Blue box detail from Supplementary Figure 1 (unit: mm). Channel depth is 20µm. The closed square and triangles designate areas for etching. (PDF 204 kb)
Rights and permissions
About this article
Cite this article
He, M., Herr, A. Automated microfluidic protein immunoblotting. Nat Protoc 5, 1844–1856 (2010). https://doi.org/10.1038/nprot.2010.142
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2010.142
This article is cited by
-
Heterotypic tumor spheroids: a platform for nanomedicine evaluation
Journal of Nanobiotechnology (2023)
-
Recent advances for cancer detection and treatment by microfluidic technology, review and update
Biological Procedures Online (2022)
-
Emerging techniques of western blotting for purification and analysis of protein
Future Journal of Pharmaceutical Sciences (2021)
-
Zinc-Finger-Protein-Based Microfluidic Electrophoretic Mobility Reversal Assay for Quantitative Double-Stranded DNA Analysis
BioChip Journal (2021)
-
Rapid Detection of Femtogram Amounts of Protein by Gel-Free Immunoblot
Bulletin of Experimental Biology and Medicine (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.