Abstract
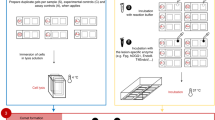
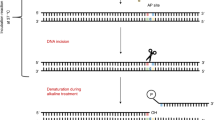
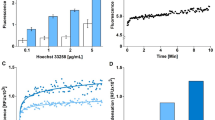
An ultrasensitive protocol is presented for the quantitative assessment of fragmented and nicked dsDNA using PicoGreen and consists of four methods. The first quantifies the concentration of DNA, whereas the second (quantitative complement of the Comet assay) quantifies the degree of DNA fragmentation seen in a typical DNA agarose electrophoresis gel. Both methods have sensitivity of 5 pg of DNA. The third method (quantitative counterpart of the electrophoresis-based qualitative apoptotic and necrotic DNA assays) quantifies the polyethylene glycol-fractionated small-size (0–1 kb) fragmented DNA. This method also detects up to 5 pg of damaged DNA and requires a minimum sample of quantity 0.2 ml of 2.5 μg ml−1. The fourth method measures the percentage of DNA nicks by alkaline DNA unwinding and requires up to 15 pg of DNA sample. The time required for processing 10 DNA samples is 1/2, 1, 13 and 1 h for the first, second, third and fourth method, respectively.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Halliwell, B. & Gutteridge, C.M.J. Free Radicals in Biology and Medicine (Oxford University Press, Oxford, 1999).
De Bont, R. & van Larebeke, N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 19, 169–185 (2004).
Van Der Vaart, M. & Pretorius, P. J. Circulating DNA: Its origin and fluctuation. Ann. N. Y. Acad. Sci. 1137, 18–26 (2008).
Gahan, P.B. Circulating nucleic acids in plasma and serum: roles in diagnosis and prognosis in diabetes and cancer. Infect. Disord. Drug Targets 8, 100–108 (2008).
Olive, P.L. & Banath, J.P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 1, 23–29 (2006).
Sambrook, J., Fritsch, F.E. & Maniatis, T. Molecular Cloning (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989).
Lelli, L.J.J., Becks, L.L., Dabrowska, I.M. & Hinshaw, B.D. ATP converts necrosis to apoptosis in oxidant-injured endothelial cells. Free Rad. Biol. Med. 25, 694–702 (1998).
Zhu, N. & Wang, Z. An assay for DNA fragmentation in apoptosis without phenol/chloroform extraction and ethanol precipitation. Anal. Biochem. 246, 155–158 (1997).
Fenech, M. The in vitro micronucleus technique. Mut. Res. 455, 81–95 (2000).
Arita, K. et al. Mechanisms of enhanced apoptosis in HL-60 cells by UV-irradiated N-3 and N-6 polyunsaturated fatty acids. Free Rad. Biol. Med. 35, 189–199 (2003).
Erusalimsky, J.D., John, J. & Moore, M. A novel filter binding assay to measure internucleosomal DNA fragmentation. Biochem. Soc. Transact. 24, 567S (1996).
Ohyama, K., Enn, P., Uchide, N., Bessho, T. & Yamakawa, T. Improvement of separation method of fragmented DNA from an apoptotic cell DNA sample for the quantitation using agarose gel electrophoresis. Biol. Pharm. Bull. 24, 342–346 (2001).
Georgiou, D.C. & Papapostolou, N. Assay for the quantification of intact/fragmented genomic DNA. Anal. Biochem. 358, 247–256 (2006).
Georgiou, D.C., Patsoukis & Papapostolou, I. Assay for the quantification of small-sized fragmented genomic DNA. Anal. Biochem. 339, 223–230 (2005).
Gavrieli, Y., Sherman, Y. & Ben-Sasson, S.A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119, 493–501 (1992).
Chohan, K.R., Griffin, J.T., Lafromboise, M., De Jonge, C.J. & Carrell, D.T. Comparison of chromatin assays for DNA fragmentation evaluation in human sperm. J. Androl. 27, 53–59 (2006).
Jones, D.P., Maellaro, E., Jiang, S., Slater, G.F.A. & Orrenius, S. Effects of N-acetyl-L-cysteine on T-cell apoptosis are not mediated by increased cellular glutathione. Immunol. Lett. 45, 205–209 (1995).
Jonas, R.C., Ziegler, R.T., Gu, H.L. & Jones, P.D. Extracellular thiol/disulfide redox state affects proliferation rate in a human colon carcinoma (CaCO2) cell line. Free Rad. Biol. Med. 33, 1499–1506 (2002).
Kanter, M.P. & Schwartz, S.H. A fluorescence enhancement assay for cellular DNA damage. Mol. Pharmacol. 22, 145–151 (1982).
Birnboim, C.H. Fluorometric analysis of DNA unwinding to study strand breaks and repair in mammalian cells. In Methods in Enzymology (eds. Packer, L. & Glazer, A.N.) 550–555 (Academic Press, San Diego, 1990).
Georgiou, C.D., Papapostolou, I., Patsoukis, N. & Grintzalis, K. Assays for the quantitative characterization of genomic, mitochondrial and plasmid DNA. In New Research on DNA Damage (eds. Kimura, H. & Suzuki, A.) (Nova Science Publishers, Inc., Hauppauge, NY, 2008) (in press).
Patsoukis, N. & Georgiou, C.D. Oxidative stress and sclerotial differentiation of the phytopathogenic fungus Rhizoctonia solani . Arch. Microbiol. 188, 225–233 (2007).
Patsoukis, N. & Georgiou, D.C. Differentiation of Sclerotinia minor depends on thiol redox state and oxidative stress. Can. J. Microbiol. 54, 28–36 (2008).
Haugland, R.P. Handbook of Fluorescent Probes and Related Products (Molecular Probes Inc., Eugene, OR, 2002).
Brown, T.A. Gene Cloning and DNA Analysis: an Introduction (Blackwell Publishing, Malden, MA, 2001).
Griffiths, A.J.F., Miller, J.H., Suzuki, D.T., Lewontin, R.C. & Gelbart, W.M. An Introduction to Genetic Analysis (W. H. Freeman, New York, NY, 2000).
Fahle, G.A. & Fischer, S.H. Comparison of six commercial DNA extraction kits for recovery of cytomegalovirus DNA from spiked human specimens. J. Clin. Microbiol. 38, 3860–3863 (2000).
Wang, L., Hirayasu, K., Ishizawa, M. & Kobayashi, Y. Purification of genomic DNA from human whole blood by isopropanol-fractionation with concentrated NaI and SDS. Nucleic Acids Res. 22, 1774–1775 (1994).
Hansen, H.M., Wiemels, J.L., Wrensch, M. & Wiencke, J.K. DNA quantification of whole genome amplified samples for genotyping on a multiplexed bead array platform. Canc. Epidemiol. Biomarkers Preven. 16, 1686–1690 (2007).
Freese, L. et al. Evaluating homogeneity of LL601 rice in commercial lots using quantitative real-time PCR. J. Agricult. Food Chem. 55, 6060–6066 (2007).
Acknowledgements
This work was financially supported by the Greek Ministry of Education, University of Patras, Greece.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Georgiou, C., Papapostolou, I. & Grintzalis, K. Protocol for the quantitative assessment of DNA concentration and damage (fragmentation and nicks). Nat Protoc 4, 125–131 (2009). https://doi.org/10.1038/nprot.2008.222
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2008.222
This article is cited by
-
Short tandem repeat (STR) instability in the oral mucosa of patients submitted to fixed orthodontic therapy: a limitation of STR profile quality for human identification
Forensic Science, Medicine and Pathology (2022)
-
A universal fluorescence-based toolkit for real-time quantification of DNA and RNA nuclease activity
Scientific Reports (2019)
-
Effect of feeding of cyclopoid copepods (Eucyclop sp.) exposed to engineered titanium dioxide nanoparticles (nTiO2) and Lead (Pb2+) on Clarias gariepinus growth and metabolism
The Journal of Basic and Applied Zoology (2018)
-
Oxidative Stress, DNA, Cell Cycle/Cell Cycle Associated Proteins and Multidrug Resistance Proteins: Targets of Human Amniotic Membrane in Hepatocellular Carcinoma
Pathology & Oncology Research (2016)
-
A new platform for ultra-high density Staphylococcus aureus transposon libraries
BMC Genomics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.