Abstract
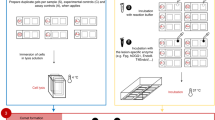
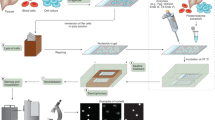
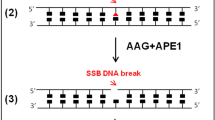
This optimized protocol (including links to instruction videos) describes a comet-based in vitro DNA repair assay that is relatively simple, versatile, and inexpensive, enabling the detection of base and nucleotide excision repair activity. Protein extracts from samples are incubated with agarose-embedded substrate nucleoids (‘naked’ supercoiled DNA) containing specifically induced DNA lesions (e.g., resulting from oxidation, UVC radiation or benzo[a]pyrene-diol epoxide treatment). DNA incisions produced during the incubation reaction are quantified as strand breaks after electrophoresis, reflecting the extract’s incision activity. The method has been applied in cell culture model systems, human biomonitoring and clinical investigations, and animal studies, using isolated blood cells and various solid tissues. Once extracts and substrates are prepared, the assay can be completed within 2 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout











Similar content being viewed by others
Data availability
The authors declare that the majority of the data shown here as examples or anticipated results are available in the original papers. Other supporting data are available upon reasonable request to the corresponding author.
References
Ostling, O. & Johanson, K. J. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 123, 291–298 (1984).
Cook, P. R., Brazell, I. A. & Jost, E. Characterization of nuclear structures containing superhelical DNA. J. Cell Sci. 22, 303–324 (1976).
Collins, A. R. & Azqueta, A. Single cell gel electrophoresis combined with lesion-specific enzymes to measure oxidative damage to DNA. in Laboratory Methods in Cell Biology, Vol. 112 (eds Anderson, C. T., Howell, E. S. & Dixit, R.) 69–92 (Elsevier, 2012).
Stefanini, M. et al. Novel Chinese hamster ultraviolet-sensitive mutants for excision repair form complementation groups 9 and 10. Cancer Res. 51, 3965–3971 (1991).
Lorenzo, Y. et al. The carotenoid beta-cryptoxanthin stimulates the repair of DNA oxidation damage in addition to acting as an antioxidant in human cells. Carcinogenesis 30, 308–314 (2009).
Collins, A. R., Fleming, I. M. & Gedik, C. M. In vitro repair of oxidative and ultraviolet-induced DNA damage in supercoiled nucleoid DNA by human cell extract. Biochim. Biophys. Acta 1219, 724–727 (1994).
Langie, S. A. et al. Development and validation of a modified comet assay to phenotypically assess nucleotide excision repair. Mutagenesis 21, 153–158 (2006).
Collins, A. R. et al. Inter-individual differences in repair of DNA base oxidation, measured in vitro with the comet assay. Mutagenesis 16, 297–301 (2001).
Møller, P. et al. Searching for assay controls for the Fpg- and hOGG1-modified comet assay. Mutagenesis 33, 9–19 (2018).
Borghini, A., Roursgaard, M., Andreassi, M. G., Kermanizadeh, A. & Moller, P. Repair activity of oxidatively damaged DNA and telomere length in human lung epithelial cells after exposure to multi-walled carbon nanotubes. Mutagenesis 32, 173–180 (2017).
Jensen, D. M. et al. Telomere length and genotoxicity in the lung of rats following intragastric exposure to food-grade titanium dioxide and vegetable carbon particles. Mutagenesis 34, 203–214 (2019).
Lohr, M. et al. Association between age and repair of oxidatively damaged DNA in human peripheral blood mononuclear cells. Mutagenesis 30, 695–700 (2015).
Gaivao, I., Piasek, A., Brevik, A., Shaposhnikov, S. & Collins, A. R. Comet assay-based methods for measuring DNA repair in vitro; estimates of inter- and intra-individual variation. Cell Biol. Toxicol. 25, 45–52 (2009).
Herrera, M. et al. Differences in repair of DNA cross-links between lymphocytes and epithelial tumor cells from colon cancer patients measured in vitro with the comet assay. Clin. Cancer Res. 15, 5466–5472 (2009).
van Dyk, E., Steenkamp, A., Koekemoer, G. & Pretorius, P. J. Hereditary tyrosinemia type 1 metabolites impair DNA excision repair pathways. Biochem. Biophys. Res. Commun. 401, 32–36 (2010).
Langie, S. A. et al. The effect of oxidative stress on nucleotide-excision repair in colon tissue of newborn piglets. Mutat. Res. 695, 75–80 (2010).
Mikkelsen, L. et al. Aging and defense against generation of 8-oxo-7,8-dihydro-2’-deoxyguanosine in DNA. Free Radic. Biol. Med. 47, 608–615 (2009).
Langie, S. A. et al. Measuring DNA repair incision activity of mouse tissue extracts towards singlet oxygen-induced DNA damage: a comet-based in vitro repair assay. Mutagenesis 26, 461–471 (2011).
Slyskova, J. et al. Functional, genetic, and epigenetic aspects of base and nucleotide excision repair in colorectal carcinomas. Clin. Cancer Res. 18, 5878–5887 (2012).
Azqueta, A., Slyskova, J., Langie, S. A., O’Neill Gaivao, I. & Collins, A. Comet assay to measure DNA repair: approach and applications. Front. Genet. 5, 288 (2014).
Yauk, C., Lambert, I., Marchetti, F. & Douglas, G. AOP 15. Alkylation of DNA in male pre-meiotic germ cells leading to heritable mutations. AOPWiki https://aopwiki.org/aops/15
Pottenger, L. H., Schoeny, R., Moore, M. & Simon, T. W. AOP 46. AFB1: mutagenic mode-of-action leading to hepatocellular carcinoma (HCC). AOPWiki https://aopwiki.org/aops/46
Silva, J. P., Gomes, A. C. & Coutinho, O. P. Oxidative DNA damage protection and repair by polyphenolic compounds in PC12 cells. Eur. J. Pharmacol. 601, 50–60 (2008).
Ramos, A. A., Azqueta, A., Pereira-Wilson, C. & Collins, A. R. Polyphenolic compounds from Salvia species protect cellular DNA from oxidation and stimulate DNA repair in cultured human cells. J. Agric. Food Chem. 58, 7465–7471 (2010).
Ramos, A. A., Pereira-Wilson, C. & Collins, A. R. Protective effects of ursolic acid and luteolin against oxidative DNA damage include enhancement of DNA repair in Caco-2 cells. Mutat. Res. 692, 6–11 (2010).
Azqueta, A., Costa, S., Lorenzo, Y., Bastani, N. E. & Collins, A. R. Vitamin C in cultured human (HeLa) cells: lack of effect on DNA protection and repair. Nutrients 5, 1200–1217 (2013).
Silva, J. P., Gomes, A. C., Proenca, F. & Coutinho, O. P. Novel nitrogen compounds enhance protection and repair of oxidative DNA damage in a neuronal cell model: comparison with quercetin. Chem. Biol. Interact. 181, 328–337 (2009).
Sliwinski, T. et al. STI571 reduces NER activity in BCR/ABL-expressing cells. Mutat. Res. 654, 162–167 (2008).
Folkmann, J. K. et al. Oxidatively damaged DNA in rats exposed by oral gavage to C60 fullerenes and single-walled carbon nanotubes. Environ. Health Perspect. 117, 703–708 (2009).
Langie, S. A. et al. Maternal folate depletion and high-fat feeding from weaning affects DNA methylation and DNA repair in brain of adult offspring. FASEB J. 27, 3323–3334 (2013).
Langie, S. A. et al. Redox and epigenetic regulation of the APE1 gene in the hippocampus of piglets: the effect of early life exposures. DNA Repair (Amst.) 18, 52–62 (2014).
Langie, S. A. et al. The ageing brain: effects on DNA repair and DNA methylation in mice. Genes (Basel) 8, https://doi.org/10.3390/genes8020075 (2017).
Setayesh, T. et al. Impact of weight loss strategies on obesity-induced DNA damage. Mol. Nutr. Food Res. 63, e1900045 (2019).
Gaivao, I. & Sierra, L. M. Drosophila comet assay: insights, uses, and future perspectives. Front. Genet. 5, 304 (2014).
Dusinska, M., Dzupinkova, Z., Wsolova, L., Harrington, V. & Collins, A. R. Possible involvement of XPA in repair of oxidative DNA damage deduced from analysis of damage, repair and genotype in a human population study. Mutagenesis 21, 205–211 (2006).
Slyskova, J. et al. Relationship between the capacity to repair 8-oxoguanine, biomarkers of genotoxicity and individual susceptibility in styrene-exposed workers. Mutat. Res. 634, 101–111 (2007).
Dusinska, M. et al. Are glutathione S transferases involved in DNA damage signalling? Interactions with DNA damage and repair revealed from molecular epidemiology studies. Mutat. Res. 736, 130–137 (2012).
Staruchova, M. et al. Occupational exposure to mineral fibres. Biomarkers of oxidative damage and antioxidant defence and associations with DNA damage and repair. Mutagenesis 23, 249–260 (2008).
Azqueta, A. et al. DNA repair as a human biomonitoring tool: comet assay approaches. Mutat. Res. 781, 71–87 (2019).
Dusinska, M. et al. Genotoxic effects of asbestos in humans. Mutat. Res. 553, 91–102 (2004).
Dusinska, M. et al. Does occupational exposure to mineral fibres cause DNA or chromosome damage? Mutat. Res. 553, 103–110 (2004).
Vodicka, P. et al. Cytogenetic markers, DNA single-strand breaks, urinary metabolites, and DNA repair rates in styrene-exposed lamination workers. Environ. Health Perspect. 112, 867–871 (2004).
Jensen, A. et al. Influence of the OGG1 Ser326Cys polymorphism on oxidatively damaged DNA and repair activity. Free Radic. Biol. Med. 52, 118–125 (2012).
Collins, A. R., Harrington, V., Drew, J. & Melvin, R. Nutritional modulation of DNA repair in a human intervention study. Carcinogenesis 24, 511–515 (2003).
Caple, F. et al. Inter-individual variation in DNA damage and base excision repair in young, healthy non-smokers: effects of dietary supplementation and genotype. Br. J. Nutr. 103, 1585–1593 (2010).
Stoyanova, E. et al. Base excision repair capacity in chronic renal failure patients undergoing hemodialysis treatment. Cell Biochem. Funct. 32, 177–182 (2014).
Fikrova, P. et al. DNA crosslinks, DNA damage and repair in peripheral blood lymphocytes of non-small cell lung cancer patients treated with platinum derivatives. Oncol. Rep. 31, 391–396 (2014).
Slyskova, J. et al. Differences in nucleotide excision repair capacity between newly diagnosed colorectal cancer patients and healthy controls. Mutagenesis 27, 225–232 (2012).
Slyskova, J. et al. Post-treatment recovery of suboptimal DNA repair capacity and gene expression levels in colorectal cancer patients. Mol. Carcinog. 54, 769–778 (2015).
Vodenkova, S. et al. Base excision repair capacity as a determinant of prognosis and therapy response in colon cancer patients. DNA Repair (Amst.) 72, 77–85 (2018).
Slyskova, J., Langie, S. A., Collins, A. R. & Vodicka, P. Functional evaluation of DNA repair in human biopsies and their relation to other cellular biomarkers. Front. Genet. 5, 116 (2014).
Singh, N. P., McCoy, M. T., Tice, R. R. & Schneider, E. L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175, 184–191 (1988).
Collins, A. R., Ord, M. J. & Johnson, R. T. Correlations of DNA damage and repair with nuclear and chromosomal damage in HeLa cells caused by methylnitrosamides. Cancer Res. 41, 5176–5187 (1981).
Crebelli, R. et al. Biomonitoring of primary aluminium industry workers: detection of micronuclei and repairable DNA lesions by alkaline SCGE. Mutat. Res. 516, 63–70 (2002).
Vande Loock, K., Decordier, I., Ciardelli, R., Haumont, D. & Kirsch-Volders, M. An aphidicolin-block nucleotide excision repair assay measuring DNA incision and repair capacity. Mutagenesis 25, 25–32 (2010).
Figueroa-Gonzalez, G. & Perez-Plasencia, C. Strategies for the evaluation of DNA damage and repair mechanisms in cancer. Oncol. Lett. 13, 3982–3988 (2017).
Lambert, B., Ringborg, U. & Skoog, L. Age-related decrease of ultraviolet light-induced DNA repair synthesis in human peripheral leukocytes. Cancer Res. 39, 2792–2795 (1979).
Athas, W. F., Hedayati, M. A., Matanoski, G. M., Farmer, E. R. & Grossman, L. Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res. 51, 5786–5793 (1991).
Redaelli, A., Magrassi, R., Bonassi, S., Abbondandolo, A. & Frosina, G. AP endonuclease activity in humans: development of a simple assay and analysis of ten normal individuals. Teratog. Carcinog. Mutagen. 18, 17–26 (1998).
Elliott, R. M., Astley, S. B., Southon, S. & Archer, D. B. Measurement of cellular repair activities for oxidative DNA damage. Free Radic. Biol. Med. 28, 1438–1446 (2000).
Roldan-Arjona, T. et al. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc. Natl Acad. Sci. USA 94, 8016–8020 (1997).
Sauvaigo, S. et al. An oligonucleotide microarray for the monitoring of repair enzyme activity toward different DNA base damage. Anal. Biochem. 333, 182–192 (2004).
Paz-Elizur, T. et al. DNA repair activity for oxidative damage and risk of lung cancer. J. Natl Cancer Inst. 95, 1312–1319 (2003).
Paz-Elizur, T. et al. Reduced repair of the oxidative 8-oxoguanine DNA damage and risk of head and neck cancer. Cancer Res. 66, 11683–11689 (2006).
Paz-Elizur, T. et al. Development of an enzymatic DNA repair assay for molecular epidemiology studies: distribution of OGG activity in healthy individuals. DNA Repair (Amst.) 6, 45–60 (2007).
Leitner-Dagan, Y. et al. N-methylpurine DNA glycosylase and OGG1 DNA repair activities: opposite associations with lung cancer risk. J. Natl Cancer Inst. 104, 1765–1769 (2012).
Leitner-Dagan, Y. et al. Enzymatic MPG DNA repair assays for two different oxidative DNA lesions reveal associations with increased lung cancer risk. Carcinogenesis 35, 2763–2770 (2014).
Azqueta, A. & Collins, A. R. The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 87, 949–968 (2013).
Riso, P. et al. DNA damage and repair activity after broccoli intake in young healthy smokers. Mutagenesis 25, 595–602 (2010).
Danielsen, P. H. et al. Oxidatively damaged DNA and its repair after experimental exposure to wood smoke in healthy humans. Mutat. Res. 642, 37–42 (2008).
Collins, A. R. & Azqueta, A. DNA repair as a biomarker in human biomonitoring studies; further applications of the comet assay. Mutat. Res. 736, 122–129 (2012).
Guarnieri, S. et al. DNA repair phenotype and dietary antioxidant supplementation. Br. J. Nutr. 99, 1018–1024 (2008).
Gaivão, I., Rodríguez, R. & Sierra, L. M. Use of the comet assay to study DNA repair in Drosophila melanogaster. in Genotoxicity and DNA Repair: A Practical Approach (eds Sierra, L. M. & Gaivão, I.) Ch. 23 (Humana Press, 2014).
Gorniak, J. P. et al. Tissue differences in BER-related incision activity and non-specific nuclease activity as measured by the comet assay. Mutagenesis 28, 673–681 (2013).
Muruzabal, D., Langie, S. A. S., Pourrut, B. & Azqueta, A. The enzyme-modified comet assay: enzyme incubation step in 2 vs 12-gels/slide systems. Mutat. Res. 845, 402981 (2019).
Azqueta, A., Langie, S. & Collins, A. R. The effect of extract concentration and time of incubation in the comet based in vitro DNA repair assay. Abstracts of the 12th International Comet Assay Workshop held at the University of Navarra, Pamplona, Spain, 29–31 August 2017. Mutagenesis 32, e24 https://academic.oup.com/mutage/article/32/6/e1/4844756#121612377 (2018).
Boer, K., Isenmann, S. & Deufel, T. Strong interference of hemoglobin concentration on CSF total protein measurement using the trichloroacetic acid precipitation method. Clin. Chem. Lab. Med. 45, 112–113 (2007).
Roman, Y., Bomsel-Demontoy, M. C., Levrier, J., Chaste-Duvernoy, D. & Jalme, M. S. Effect of hemolysis on plasma protein levels and plasma electrophoresis in birds. J. Wildl. Dis. 45, 73–80 (2009).
Brodersen, R. Bilirubin. Solubility and interaction with albumin and phospholipid. J. Biol. Chem. 254, 2364–2369 (1979).
Kjellin, K. G. Bilirubin compounds in the CSF. J. Neurol. Sci. 13, 161–173 (1971).
Moller, P. et al. On the search for an intelligible comet assay descriptor. Front. Genet. 5, 217 (2014).
Forchhammer, L. et al. Variation in assessment of oxidatively damaged DNA in mononuclear blood cells by the comet assay with visual scoring. Mutagenesis 23, 223–231 (2008).
Azqueta, A. et al. The influence of scoring method on variability in results obtained with the comet assay. Mutagenesis 26, 393–399 (2011).
Brunborg, G., Rolstadaas, L. & Gutzkow, K. B. Electrophoresis in the comet assay. in Electrophoresis: Life Sciences Practical Applications (eds Boldura, O-M. & Baltă, C.) 526–652 (IntechOpen, 2018).
Shaposhnikov, S. et al. Twelve-gel slide format optimised for comet assay and fluorescent in situ hybridisation. Toxicol. Lett. 195, 31–34 (2010).
Collins, A. R. et al. The comet assay: topical issues. Mutagenesis 23, 143–151 (2008).
Forchhammer, L. et al. Variation in the measurement of DNA damage by comet assay measured by the ECVAG inter-laboratory validation trial. Mutagenesis 25, 113–123 (2010).
Azqueta, A., Langie, S. A., Slyskova, J. & Collins, A. R. Measurement of DNA base and nucleotide excision repair activities in mammalian cells and tissues using the comet assay—a methodological overview. DNA Repair (Amst.) 12, 1007–1010 (2013).
Gungor, N. et al. Lung inflammation is associated with reduced pulmonary nucleotide excision repair in vivo. Mutagenesis 25, 77–82 (2010).
Azqueta, A. et al. A comparative performance test of standard, medium- and high-throughput comet assays. Toxicol. Vitr. 27, 768–773 (2013).
Brauner, E. V. et al. Exposure to ultrafine particles from ambient air and oxidative stress-induced DNA damage. Environ. Health Perspect. 115, 1177–1182 (2007).
Moller, P. et al. Measurement of oxidative damage to DNA in nanomaterial exposed cells and animals. Environ. Mol. Mutagen. 56, 97–110 (2015).
Hasplova, K. et al. DNA alkylation lesions and their repair in human cells: modification of the comet assay with 3-methyladenine DNA glycosylase (AlkD). Toxicol. Lett. 208, 76–81 (2012).
Dusinska, M. et al. Testing strategies for the safety of nanoparticles used in medical applications. Nanomed. (Lond.) 4, 605–607 (2009).
Choi, S. W., Yeung, V. T., Collins, A. R. & Benzie, I. F. Redox-linked effects of green tea on DNA damage and repair, and influence of microsatellite polymorphism in HMOX-1: results of a human intervention trial. Mutagenesis 30, 129–137 (2015).
Brevik, A. et al. Supplementation of a Western diet with golden kiwifruits (Actinidia chinensis var.‘Hort 16A’): effects on biomarkers of oxidation damage and antioxidant protection. Nutr. J. 10, 54 (2011).
Hanova, M. et al. Modulation of DNA repair capacity and mRNA expression levels of XRCC1, hOGG1 and XPC genes in styrene-exposed workers. Toxicol. Appl. Pharmacol. 248, 194–200 (2010).
Humphreys, V. et al. Age-related increases in DNA repair and antioxidant protection: a comparison of the Boyd Orr Cohort of elderly subjects with a younger population sample. Age Ageing 36, 521–526 (2007).
Langie, S. A. et al. Modulation of nucleotide excision repair in human lymphocytes by genetic and dietary factors. Br. J. Nutr. 103, 490–501 (2010).
Al-Serori, H. et al. Mobile phone specific electromagnetic fields induce transient DNA damage and nucleotide excision repair in serum-deprived human glioblastoma cells. PLoS One 13, e0193677 (2018).
Soares, J. P. et al. Effects of combined physical exercise training on DNA damage and repair capacity: role of oxidative stress changes. Age (Dordr.) 37, 9799 (2015).
Acknowledgements
We thank the hCOMET project (COST Action, CA 15132) for support. A.A. thanks the Ministry of Economy, Industry and Competitiveness (‘Ramón y Cajal’ programme, RYC2013-14370) of the Spanish Government for personal support. P.V. acknowledges support from the National Science Foundation (19-10543S).
Author information
Authors and Affiliations
Contributions
S.V., A.A., R.W.L.G. and S.A.S.L. designed figures; S.A.S.L. provided anticipated results and managed the manuscript preparation; S.V., A.A. and S.A.S.L. drafted the paper; and A.C., M.D., P.M. A.O., I.G. and P.V. contributed to and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Ricard Marcos and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Related links
Key references using this protocol
Gaivão, I., Piasek, A., Brevik, A., Shaposhnikov, S. & Collins, A. R. Cell Biol. Toxicol. 25, 45–52 (2009): https://doi.org/10.1007/s10565-007-9047-5
Langie, S. A. et al. Mutagenesis 26, 461–471 (2011): https://doi.org/10.1093/mutage/ger005
Slyskova, J. et al. Clin. Cancer Res. 18, 5878–5887 (2012): https://doi.org/10.1158/1078-0432.CCR-12-1380
Supplementary information
Rights and permissions
About this article
Cite this article
Vodenkova, S., Azqueta, A., Collins, A. et al. An optimized comet-based in vitro DNA repair assay to assess base and nucleotide excision repair activity. Nat Protoc 15, 3844–3878 (2020). https://doi.org/10.1038/s41596-020-0401-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-0401-x
This article is cited by
-
Comparison of comet-based approaches to assess base excision repair
Archives of Toxicology (2023)
-
Measuring DNA modifications with the comet assay: a compendium of protocols
Nature Protocols (2023)
-
Hedyotis diffusae Herba-Andrographis Herba inhibits the cellular proliferation of nasopharyngeal carcinoma and triggers DNA damage through activation of p53 and p21
Cancer Gene Therapy (2022)
-
Cell survival after DNA damage in the comet assay
Archives of Toxicology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.