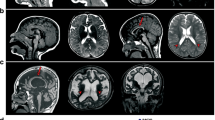
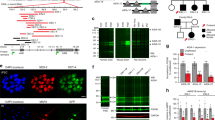
Abstract
The NONO protein has been characterized as an important transcriptional regulator in diverse cellular contexts. Here we show that loss of NONO function is a likely cause of human intellectual disability and that NONO-deficient mice have cognitive and affective deficits. Correspondingly, we find specific defects at inhibitory synapses, where NONO regulates synaptic transcription and gephyrin scaffold structure. Our data identify NONO as a possible neurodevelopmental disease gene and highlight the key role of the DBHS protein family in functional organization of GABAergic synapses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Referenced accessions
NCBI Reference Sequence
References
Fox, A.H., Bond, C.S. & Lamond, A.I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell 16, 5304–5315 (2005).
Myojin, R. et al. Expression and functional significance of mouse paraspeckle protein 1 on spermatogenesis. Biol. Reprod. 71, 926–932 (2004).
Shav-Tal, Y. & Zipori, D. PSF and p54nrb/NonO – multi-functional nuclear proteins. FEBS Lett. 531, 109–114 (2002).
Dong, X., Sweet, J., Challis, J.R., Brown, T. & Lye, S.J. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol. Cell. Biol. 27, 4863–4875 (2007).
Amelio, A.L. et al. A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proc. Natl. Acad. Sci. USA 104, 20314–20319 (2007).
Park, Y., Lee, J.M., Hwang, M.Y., Son, G.H. & Geum, D. NonO binds to the CpG island of oct4 promoter and functions as a transcriptional activator of oct4 gene expression. Mol. Cells 35, 61–69 (2013).
Kim, K.K., Kim, Y.C., Adelstein, R.S. & Kawamoto, S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 39, 3064–3078 (2011).
Patton, J.G., Porro, E.B., Galceran, J., Tempst, P. & Nadal-Ginard, B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 7, 393–406 (1993).
Kaneko, S., Rozenblatt-Rosen, O., Meyerson, M. & Manley, J.L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev. 21, 1779–1789 (2007).
Kanai, Y., Dohmae, N. & Hirokawa, N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 (2004).
Izumi, H., McCloskey, A., Shinmyozu, K. & Ohno, M. p54nrb/NonO and PSF promote U snRNA nuclear export by accelerating its export complex assembly. Nucleic Acids Res. 42, 3998–4007 (2014).
Nakagawa, S. & Hirose, T. Paraspeckle nuclear bodies—useful uselessness? Cell. Mol. Life Sci. 69, 3027–3036 (2012).
Bond, C.S. & Fox, A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 (2009).
Chen, L.L., DeCerbo, J.N. & Carmichael, G.G. Alu element-mediated gene silencing. EMBO J. 27, 1694–1705 (2008).
Prasanth, K.V. et al. Regulating gene expression through RNA nuclear retention. Cell 123, 249–263 (2005).
Zhang, Z. & Carmichael, G.G. The fate of dsRNA in the nucleus: a p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106, 465–475 (2001).
Brown, S.A. et al. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308, 693–696 (2005).
Duong, H.A., Robles, M.S., Knutti, D. & Weitz, C.J. A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439 (2011).
Kowalska, E. et al. NONO couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. USA 110, 1592–1599 (2013).
Kowalska, E. et al. Distinct roles of DBHS family members in the circadian transcriptional feedback loop. Mol. Cell. Biol. 32, 4585–4594 (2012).
de Ligt, J. et al. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 367, 1921–1929 (2012).
Rauch, A. et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 380, 1674–1682 (2012).
Vissers, L.E., de Vries, B.B. & Veltman, J.A. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J. Med. Genet. 47, 289–297 (2010).
Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature 519, 223–228 (2015).
Tyagarajan, S.K. & Fritschy, J.M. Gephyrin: a master regulator of neuronal function? Nat. Rev. Neurosci. 15, 141–156 (2014).
Tyagarajan, S.K. et al. Regulation of GABAergic synapse formation and plasticity by GSK3β-dependent phosphorylation of gephyrin. Proc. Natl. Acad. Sci. USA 108, 379–384 (2011).
Panzanelli, P. et al. Distinct mechanisms regulate GABAA receptor and gephyrin clustering at perisomatic and axo-axonic synapses on CA1 pyramidal cells. J. Physiol. (Lond.) 589, 4959–4980 (2011).
Jacob, T.C. et al. Gephyrin regulates the cell surface dynamics of synaptic GABAA receptors. J. Neurosci. 25, 10469–10478 (2005).
Mullen, S.A. et al. Copy number variants are frequent in genetic generalized epilepsy with intellectual disability. Neurology 81, 1507–1514 (2013).
Zhang, G., Neubert, T.A. & Jordan, B.A. RNA binding proteins accumulate at the postsynaptic density with synaptic activity. J. Neurosci. 32, 599–609 (2012).
Ruby, N.F. et al. Hippocampal-dependent learning requires a functional circadian system. Proc. Natl. Acad. Sci. USA 105, 15593–15598 (2008).
Castner, S.A. et al. Reversal of ketamine-induced working memory impairments by the GABAAalpha2/3 agonist TPA023. Biol. Psychiatry 67, 998–1001 (2010).
Hofmann, M. et al. Assessment of the effects of NS11394 and L-838417, alpha2/3 subunit-selective GABA(A) [corrected] receptor-positive allosteric modulators, in tests for pain, anxiety, memory and motor function. Behav. Pharmacol. 23, 790–801 (2012).
Hines, R.M. et al. Disrupting the clustering of GABAA receptor α2 subunits in the frontal cortex leads to reduced gamma-power and cognitive deficits. Proc. Natl. Acad. Sci. USA 110, 16628–16633 (2013).
Lehner, M. et al. Differences in the density of GABA-A receptor alpha-2 subunits and gephyrin in brain structures of rats selected for low and high anxiety in basal and fear-stimulated conditions, in a model of contextual fear conditioning. Neurobiol. Learn. Mem. 94, 499–508 (2010).
Yang, B. et al. Sh3pxd2b mice are a model for craniofacial dysmorphology and otitis media. PLoS ONE 6, e22622 (2011).
Baltes, C., Radzwill, N., Bosshard, S., Marek, D. & Rudin, M. Micro MRI of the mouse brain using a novel 400 MHz cryogenic quadrature RF probe. NMR Biomed. 22, 834–842 (2009).
Mohajeri, M.H. et al. Intact spatial memory in mice with seizure-induced partial loss of hippocampal pyramidal neurons. Neurobiol. Dis. 12, 174–181 (2003).
Gallagher, M., Burwell, R. & Burchinal, M. Severity of spatial learning impairment in aging: development of a learning index for performance in the Morris water maze. Behav. Neurosci. 107, 618–626 (1993).
Whishaw, I.Q. Cholinergic receptor blockade in the rat impairs locale but not taxon strategies for place navigation in a swimming pool. Behav. Neurosci. 99, 979–1005 (1985).
Cros, J. et al. Human CD14dim monocytes patrol and sense nucleic acids and viruses via TLR7 and TLR8 receptors. Immunity 33, 375–386 (2010).
Schmittgen, T.D. & Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 (2008).
Li, B. & Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Buerli, T. et al. Efficient transfection of DNA or shRNA vectors into neurons using magnetofection. Nat. Protoc. 2, 3090–3101 (2007).
Lardi-Studler, B. et al. Vertebrate-specific sequences in the gephyrin E-domain regulate cytosolic aggregation and postsynaptic clustering. J. Cell Sci. 120, 1371–1382 (2007).
Kuwahara, S. et al. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol. Reprod. 75, 352–359 (2006).
Fritschy, J.M. & Mohler, H. GABAA-receptor heterogeneity in the adult rat brain: differential regional and cellular distribution of seven major subunits. J. Comp. Neurol. 359, 154–194 (1995).
Brunig, I., Suter, A., Knuesel, I., Luscher, B. & Fritschy, J.M. GABAergic terminals are required for postsynaptic clustering of dystrophin but not of GABAA receptors and gephyrin. J. Neurosci. 22, 4805–4813 (2002).
Notter, T., Panzanelli, P., Pfister, S., Mircsof, D. & Fritschy, J.M. A protocol for concurrent high-quality immunohistochemical and biochemical analyses in adult mouse central nervous system. Eur. J. Neurosci. 39, 165–175 (2014).
Dunkley, P.R., Jarvie, P.E. & Robinson, P.J. A rapid Percoll gradient procedure for preparation of synaptosomes. Nat. Protoc. 3, 1718–1728 (2008).
Acknowledgements
We are grateful to the patients and their family members for their participation in our study and to the Functional Genomics Center Zürich (FGCZ) for transcriptomic services. This program has received a state subsidy managed by the National Research Agency under the Investments for the Future program bearing the reference ANR-10-IAHU-01.This study was also supported by the Centre National de la Recherche Scientifique (CNRS), the Fondation pour la Recherche Médicale (DEQ20120323702) and the Ministère de la Recherche et de l'Enseignement Supérieur, as well as by the Swiss National Science Foundation, the Zurich Clinical Research Priority Program “Sleep and Health,” the Zurich Fonds zur Förderung des akademischen Nachwuchses and the Zurich Neurozentrum (ZNZ). D.M., S.A.B. and S.K.T. are affiliates of the ZNZ Life Sciences Zurich graduate program, and S.A.B. is a member of the Zürich Center for Interdisciplinary Sleep Research (ZIS). The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the UK Department of Health, and the Wellcome Trust Sanger Institute (grant number WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The research team acknowledges the support of the UK National Institute for Health Research, through the Comprehensive Clinical Research Network.
Author information
Authors and Affiliations
Consortia
Contributions
L.C., S.A.B., J.-M.F. and S.K.T. designed the study. J.A., M. Rio and K.P. recruited and evaluated the study subjects. N.B. performed and analyzed the human brain imaging, and D.M., P.S., A.S. and M. Rudin performed and analyzed the mouse brain imaging. M.L. analyzed whole-exome sequencing data and performed transcriptional analysis and western blot analysis on patient cells, and L.G. performed circadian analyses on patient cells. S.M. contributed to whole-exome sequencing data analysis. C.B.-F. performed whole-exome sequencing. N.C. and P.N. performed the bioinformatics studies. R.H. and S.J.M. supplied engineered virus for in vivo rescue experiments. D.M. executed and analyzed all experiments on mouse tissues, and D.M. and M.Ž. executed and analyzed experiments in rodent cells. D.M., C.K., F.C., A.-K.F. and D.P.W. performed and analyzed mouse behavioral experiments. A.M. contributed to the clinical evaluation of the patients. K.S.-P. and O.A. contributed to the WES analysis. G.B. prepared cultured neurons for all experiments in this study. All authors contributed to writing and editing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A full list of consortium members appears in the Supplementary Note.
Integrated supplementary information
Supplementary Figure 1 Molecular analyses of patients MCCID1 and MCCID2
(a) Sanger sequencing chromatograms showing the NONO mutations in the probands and their parents. (b) Schematic representation of the NONO transcript showing exon structure. (c) Schematic overview of the NONO protein showing the different functional domains.
Supplementary Figure 3 Full-length western blot of NONO
Western blot of NONO protein in patients and controls, using a different polyclonal antibody from the one used in Figure 1.
Supplementary Figure 4 Reduced circadian amplitude in patient fibroblasts.
(a) Graph of average circadian oscillations of bioluminescence from Bmal1-luciferase reporter-infected fibroblasts from patients (grey) or siblings (black). Y-axis, background-subtracted bioluminescence in photons per minute; X-axis, time in days. Depicted curve is the average of three independent experiments in technical quadruplicate. (b) Circadian period measured in cells from controls (black bars) and patients (open bars). (c) Circadian amplitude measured in the same cells (arbitrary units). Student t-test, p<0.001. Values for both panels are the average of three independent experiments with fibroblasts from both patients and their siblings in technical quadruplicate.
Supplementary Figure 5 Brain morphology in Nonogt mice.
(a) Photograph of representative brains from WT (left) and Nonogt (right) mice. (b) Quantification of weight of whole brain, cortex, and cerebellum. N=5-9 mice per genotype. Student t-test. White bars, WT. grey bars, Nonogt. In this and subsequent figures, ***p<0.001, **p<0.01, *p<0.05. Quantification of all brain parameters from different morphological tests is shown in detail in Table S4.
Supplementary Figure 6 Anxiety-related phenotypes in Nonogt mice.
(a) Percentage of time spent in open arms of elevated plus-maze. N=13-14 mice per genotype. Student t-test, p<0.001 (b) Post-conditioning startle response in prepulse inhibition test. Y-axis, whole-body startle response in volts; X-axis, stimulus in decibels. N=13-14 mice per genotype. Student t-test, p<0.01 or 0.001 as indicated. (c) Open-field exploration. Y-axis, percentage of area surface tiles visited. X-axis, subsequent 5-minute intervals after commencement of test. N=18 mice per genotype, repeated ANOVA, gene p<.0002, time p<.0001, time x gene n.s. (d) Light-dark transition test. Y-axis, percentage of time spent in zones indicated on X-axis. N=18 mice per genotype, repeated ANOVA, gene p<.0001, zone p<.0001, zone x gene p<0.0001. In all panels, Nonogt mice are represented by grey bars/circles, compared to WT littermates (open).
Supplementary Figure 7 Cell-type- and layer-specific localization of NONO in mouse brain.
(a) Staining of hippocampal cell nuclei by DAPI (blue), anti-NONO (green), and neuron-specific anti-NeuN or astrocyte-specific anti-GFAP (red, left or right column respectively). (b) Identical staining of cortex.
Supplementary Figure 8 Widespread dysregulation of transcription in Nonogt mouse hippocampi.
(a) Volcano plot of deregulated genes. Red dots, p<0.01 and log2>0.5. (b) Reduced GABRA2 mRNA levels in hippocampi of Nonogt mice. White bars, WT. grey bars, Nonogt. Student t-test, N=4. (c) List of most severely deregulated transcripts, showing upregulation of sister DBHS family members Sfpq and Pspc1.
Supplementary Figure 9 Overexpression of PSPC1 and SFPQ protein in Nonogt mouse hippocampi.
(a) Western blots of hippocampal protein extracts from widtype and Nonogt mice hippocampi, probed with anti-NONO, anti-PSPC1, anti-SFPQ, and anti-β-actin. N=3 mice per genotype. (b) Quantification of (a). White bars, WT. Black bars, Nonogt. Student t-test, p<0.01.
Supplementary Figure 10 Transcript abundance in different neuronal compartments in Nonogt mice.
(a) Profile of the mouse forebrain synaptic transcriptome. Y-axis, ratio of transcript reads from synaptome RNA sequencing compared to total. Selected transcripts previously characterized to be transported to synapses (Kif5a, Shank3, CamK2A, Arc, Gabra2), present throughout the cell (Map2, Actb), or retained in the nucleus (Neat1) are indicated to demonstrate the quality of synaptosomal transcript enrichment. Blue shading, transcripts more than 1.5x enriched. (b) Quantification of Neat1 compared to Gapdh in whole-cell homogenate, purified nuclei, supernatant, or gradient-purified synaptosomes from WT (white bars) or Nonogt mice (solid bars). (c) Quantification of CamkII compared to Gapdh. (d) Quantification of Gabra2 compared to Gapdh.
Supplementary Figure 11 Reduction of synaptic GABRA2 in Nonogt mice.
(a) Western blot showing fractionation of mouse forebrain into different neuronal compartments by density gradient centrifugation. Immunohistochemistry using antibodies against NONO, gephyrin, and GABA is pictured. As a control, β-actin and dendrite-enriched PSD-95 are also shown. (b) Quantification of the reduction in GABRA2 levels in each compartment.
Supplementary Figure 12 Specific reduction of Gabra2 transcript levels in cultured Nonogt neurons.
(a) Single-transcript-resolution RNA in-situ hybridization to detect abundance of collybistin (CB, blue), Gabra1 (blue), Gabra2 (red), and CamKII (red) in WT or Nonogt neurons. Two transcripts were tested in different colors in a single plate of cells, and depicted in a single column. (b) Quantification of collybistin transcript distribution in puncta number per cell for 44-56 cells from 2 experiments. (C) Similar quantification of Gabra2. Solid circles, Nonogt neurons; open circles, WT.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Tables 1–7 and Supplementary Note (PDF 3997 kb)
Source data
Rights and permissions
About this article
Cite this article
Mircsof, D., Langouët, M., Rio, M. et al. Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nat Neurosci 18, 1731–1736 (2015). https://doi.org/10.1038/nn.4169
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nn.4169
This article is cited by
-
Liquid-liquid phase separation in DNA double-strand breaks repair
Cell Death & Disease (2023)
-
A novel NONO variant that causes developmental delay and cardiac phenotypes
Scientific Reports (2023)
-
Nono deficiency impedes the proliferation and adhesion of H9c2 cardiomyocytes through Pi3k/Akt signaling pathway
Scientific Reports (2023)
-
Molecular signature of excessive female aggression: study of stressed mice with genetic inactivation of neuronal serotonin synthesis
Journal of Neural Transmission (2023)
-
Human ARHGEF9 intellectual disability syndrome is phenocopied by a mutation that disrupts collybistin binding to the GABAA receptor α2 subunit
Molecular Psychiatry (2022)