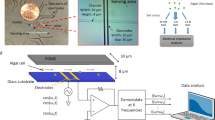
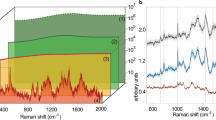
Abstract
Understanding metabolism in live microalgae is crucial for efficient biomaterial engineering, but conventional methods fail to evaluate heterogeneous populations of motile microalgae due to the labelling requirements and limited imaging speeds. Here, we demonstrate label-free video-rate metabolite imaging of live Euglena gracilis and statistical analysis of intracellular metabolite distributions under different culture conditions. Our approach provides further insights into understanding microalgal heterogeneity, optimizing culture methods and screening mutant microalgae.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Georgianna, D. R. & Mayfield, S. P. Nature 488, 329–335 (2012).
Wijffels, R. H. & Barbosa, M. J. Science 329, 796–799 (2010).
Harun, R. et al. Renew. Sustain. Energy Rev. 14, 1037–1047 (2010).
Buetow, D. E. The Biology of Euglena Vol. 9, Ch. 1 (Academic, 1989).
Koizumi, N. et al. Antiviral Res. 21, 1–14 (1993).
Watanabe, T. et al. Food Funct. 4, 1685–1690 (2013).
Demirbas, A. & Demirbas, M. F. Energ. Convers. Manage. 52, 163–170 (2011).
Altschuler, S. J. & Wu, L. F. Cell 141, 559–563 (2010).
Lisec, J. et al. Nature Protoc. 1, 387–396 (2006).
Graham, M. D. J. Lab. Autom. 8, 72–81 (2003).
Bonner, W. A. et al. Rev. Sci. Instrum. 43, 404–409 (1972).
Lichtman, J. W. & Conchello, J.-A. Nature Methods 2, 910–919 (2005).
Lakowicz, J. R. Principles of Fluorescence Spectroscopy (Springer, 2006).
Freudiger, C. W. et al. Science 322, 1857–1861 (2008).
Cheng, J.-X. & Xie, X. S. Science 350, aaa8870 (2015).
Ozeki, Y. et al. Nature Photon. 6, 845–851 (2012).
Littlejohn, G. R. et al. Plant Physiol. 168, 18–28 (2015).
He, X. N. et al. Biomed. Opt. Express 3, 2896–2906 (2012).
Cavonius, L. et al. Plant Physiol. 167, 603–616 (2015).
Coleman, L. W. et al. Plant. Cell Physiol. 29, 423–432 (1988).
Acknowledgements
This work was funded mainly by the ImPACT Program of the Council for Science, Technology and Innovation (Cabinet Office, Government of Japan) and partly by Advanced Photon Science Alliance and the Japan Society for the Promotion of Science (JSPS) KAKENHI (grant no. 25702026). Y.S. and K.G. are supported by JSPS and partly by Burroughs Wellcome Fund, respectively.
Author information
Authors and Affiliations
Contributions
Y.W. and Y.S. performed the experiments. O.I., A.N. and T.I. prepared the samples. Y.W., Y.S., M.H., R.D., M.S., N.T., T.S. and H.W. performed the data analysis. Y.W., Y.S., K.G. and Y.O. wrote the manuscript. K.G. conceived the concept. K.S., K.G. and Y.O. supervised the work.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Discussion, Supplementary References, Supplementary Figures 1–8, Legends for Supplementary Videos 1 and 2. (PDF 1993 kb)
Supplementary Video 1
Comparison in SRS imaging of motile E. gracilis in motion between frame rates of 27 fps (top) and 6.75 fps (bottom). (AVI 3246 kb)
Supplementary Video 1
3D metabolite imaging of E. gracilis. (MOV 2134 kb)
Rights and permissions
About this article
Cite this article
Wakisaka, Y., Suzuki, Y., Iwata, O. et al. Probing the metabolic heterogeneity of live Euglena gracilis with stimulated Raman scattering microscopy. Nat Microbiol 1, 16124 (2016). https://doi.org/10.1038/nmicrobiol.2016.124
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/nmicrobiol.2016.124
This article is cited by
-
Photoswitchable polyynes for multiplexed stimulated Raman scattering microscopy with reversible light control
Nature Communications (2024)
-
Fast Real-Time Brain Tumor Detection Based on Stimulated Raman Histology and Self-Supervised Deep Learning Model
Journal of Imaging Informatics in Medicine (2024)
-
Computational coherent Raman scattering imaging: breaking physical barriers by fusion of advanced instrumentation and data science
eLight (2023)
-
Label-free mid-infrared photothermal live-cell imaging beyond video rate
Light: Science & Applications (2023)
-
Instant diagnosis of gastroscopic biopsy via deep-learned single-shot femtosecond stimulated Raman histology
Nature Communications (2022)