Abstract
To determine the correlation between BRAF genotype and MLH1 promoter methylation in a screening program for Lynch syndrome (LS), a universal screening program for LS was established in two medical centers. Tumors with abnormal MLH1 staining were evaluated for both BRAF V600E genotype and MLH1 promoter methylation. Tumors positive for both were considered sporadic, and genetic testing was recommended for all others. A total 1011 colorectal cancer cases were screened for Lynch syndrome, and 148 (14.6%) exhibited absent MLH1 immunostaining. Both BRAF and MLH1 methylation testing were completed in 126 cases. Concordant results (both positive or both negative) were obtained in 86 (68.3%) and 16 (12.7%) cases, respectively, with 81% concordance overall. The positive and negative predictive values for a BRAF mutation in predicting MLH1 promoter methylation were 98.9% and 41%, respectively, and the negative predictive value fell to 15% in patients ≥70 years old. Using BRAF genotyping as a sole test to evaluate cases with absent MLH1 staining would have increased referral rates for genetic testing by 2.3-fold compared with MLH1 methylation testing alone (31% vs 13.5%, respectively, P<0.01). However, a hybrid approach that reserves MLH1 methylation testing for BRAF wild-type cases only would significantly decrease the number of methylation assays performed and reduce the referral rate for genetic testing to 12.7%. A BRAF mutation has an excellent positive predictive value but poor negative predictive value in predicting MLH1 promoter methylation. A hybrid use of these tests may reduce the number of low-risk patients referred to genetic counseling and facilitate wider implementation of Lynch syndrome screening programs.
Similar content being viewed by others
Main
Colorectal cancer remains the second leading cause of cancer-related mortality affecting both men and women in the United States and Europe.1, 2, 3 Most colorectal cancer cases are considered to be sporadic. Approximately 5% of cases arise in the context of a hereditary colon cancer syndrome, and Lynch syndrome is the most common of these conditions.4 Unfortunately, Lynch syndrome is under-recognized in the population.
Individuals and families with Lynch syndrome require enhanced cancer screening as well as more aggressive surgical approaches if cancer is diagnosed. An evaluation for Lynch syndrome has traditionally been triggered by clinical criteria (eg, Amsterdam and Revised Bethesda criteria).4 However, the practical application of these criteria is limited by their complexity and accuracy of a self-reported family history. This has led to an endorsement for universal screening, in which all the colorectal tumors are evaluated for Lynch syndrome.4, 5, 6
The current approach to screening for Lynch syndrome includes tumor testing with immunohistochemistry for the four DNA mismatch repair proteins that underlie Lynch syndrome or testing tumor DNA for microsatellite instability. Immunohistochemistry is preferred due to the lower cost, wider availability, and the ability to direct the subsequent genetic evaluation.5, 7, 8 Specifically, germline genetic testing is recommended when abnormal MSH-2, MSH-6, or PMS-2 staining is observed. However, the most challenging finding in universal screening programs is the evaluation of abnormal MLH1 staining. The vast majority of cases with MLH1 loss are not due to a germline mutation in the MLH1 gene but rather somatic methylation of the MLH1 gene promoter. Approximately 12% of colorectal cancers exhibit MLH1 promoter methylation.9
There are currently two assays to determine whether MLH1 promoter methylation is present and thereby obviate the need for blood genetic testing for Lynch syndrome. The first is a direct assay for MLH1 promoter methylation that is performed on tumor DNA.7, 8, 10 BRAF testing for the V600E mutation is an alternative molecular approach to distinguish between sporadic and Lynch syndrome-associated colorectal cancer cases.10, 11 BRAF testing has become routine in the evaluation of colorectal cancer and is technically less demanding and less costly than assays of MLH1 promoter methylation.12 However, the correlations between BRAF genotype and MLH1 promoter methylation have not been well defined in the screening population.13, 14 This is reflected in the wide variability of recommendations for evaluation of loss of MLH1 staining that include BRAF testing, MLH1 promoter methylation testing, or a combination of both BRAF and MLH1 promoter methylation testing.4, 15 Currently, there is a knowledge gap in the performance characteristics of these approaches in a large screening population.11 As universal screening strategies for Lynch syndrome become more widespread, understanding the relationship between BRAF and MLH1 promoter methylation testing has important implications for cost and accuracy. An optimized screening strategy will reduce the number of low-risk patients referred to genetic counseling while maximizing the detection of Lynch syndrome in the population.
The aim of this study is to characterize the correlation between BRAF V600E genotype and MLH1 promoter methylation in the setting of a universal screening program for Lynch syndrome.
Materials and methods
Study Group and Design
A universal Lynch syndrome screening program was implemented for all newly diagnosed colorectal adenocarcinomas at two sites: Massachusetts General Hospital, a tertiary care center in Boston, MA, and North Shore Medical Center, a community hospital in Danvers, MA. Immunohistochemical staining for mismatch repair proteins was performed on all colorectal tumors collected during a 4-year period (January 2011–December 2014). Tumors with intact immunohistochemical staining were considered sporadic, and further risk assessment for Lynch syndrome was not indicated.
In cases where abnormal MLH1 staining was demonstrated (defined as absent or very faint staining, which may also occur in LS16), BRAF V600E mutation analysis and MLH1 promoter methylation testing were performed. Among patients with abnormal MLH1 staining, patients with double-positive tests (BRAF mutation positive and MLH1 promoter methylation positive) were considered to have sporadic tumors and no further genetic testing was performed (group A). In patients with double-negative results (BRAF wild type and no MLH1 promoter methylation, designated group B) and patients with discordant BRAF mutation and MLH1 promoter methylation results (group C), a recommendation for genetic counseling for discussion of germline genetic testing was made. This recommendation was conveyed to the patient’s primary clinician.
A chart review was performed in all the cases with loss of MLH1 staining. Medical history and histological finding were recorded. The study was approved by the institutional IRBs.
Immunohistochemical Staining for Mismatch Repair Proteins
Immunohistochemical testing was performed using automated staining technique (Leica Bond III and Ventana Benchmark Ultra platform at the Massachusetts General Hospital and North Shore Medical Center, respectively).
For specimens from the North Shore Medical Center, the clones of the MLH1, MSH2, MSH6, and PMS6 pre-diluted antibodies (Ventana) were M1, G219-1129, 44, and EPR3947 respectively. For specimens from the Massachusetts General Hospital, the clones (and dilutions) of antibodies for MLH1, MSH2, and MSH6 (Biocare Medical) were CM220C (1:50), CM219C (1:25), and CM265C(1:25), respectively. The anti PMS2 clone (BD Biosciences) was 556415 (diluted 1:100). In each center, these tests were performed as per standard clinical protocols and described below.
Five micrometer-thick consecutive sections were deparaffinized and hydrated through a graded series of alcohols. Antigen retrieval was performed with 10 mmol/l citrate buffer (pH 6.0) in a microwave oven for 10 min, after which endogenous peroxidase activity was inhibited by immersion in 3% hydrogen peroxide/methanol solution. The sections then were incubated with the primary antibodies followed by thorough washing in phosphate-buffered solution. Next, they were incubated with the biotinylated secondary antibody and then with an avidin-biotinylated horseradish peroxidase complex (Vectastain Elite ABC kit; Vector Laboratories, Burlingame, CA, USA), after which they were developed by using 3,3′-diaminobenzidine tetrachloride as the chromogen. Nuclear counterstaining was achieved with Mayer’s hematoxylin.
Protein staining was interpreted as intact expression (normal) if there was any positive reaction in the tumor cell nuclei. An interpretation of loss of expression (negative) was made when there was no staining of tumor cell nuclei while a positive reaction was present in internal control cell nuclei, such as stromal, inflammatory, or non-neoplastic epithelial cells.
BRAF Mutation Testing
Mutational analysis of BRAF was restricted to the V600E alteration. For patients evaluated at the Massachusetts General Hospital, BRAF mutations were detected from fixed formalin paraffin embedded DNA using the Snapshot mutation detection platform as previously described.17 The samples from the North Shore Medical Center were sent to a referral laboratory (Clarient Diagnostic Services Inc, Aliso Viejo, CA) using a lab developed allele-specific polymerase chain reaction (PCR)-based assay. The tumor area on an H&E slide for this formalin fixed paraffin embedded specimen was identified by a pathologist. Tissue from the corresponding sections mounted on unstained slides was lysed and the genomic DNA was purified from the sample. Real-time PCR amplification and detection of target DNA (approximately 100-base pair sequence of human DNA containing the BRAF codon 600 site in exon 15) using a complementary primer pair and two oligonucleotide probes labeled with different fluorescent dyes was performed. In the PCR reaction, BRAF wild-type and mutant DNA target-specific, fluorescent dye-labeled TaqMan probes bind to the wild-type and mutant sequences, respectively. Two independent duplex reactions per patient sample are performed. Kinetic analysis of the fluorescent signals reveals the presence of potential BRAF mutations. Three external run controls (positive, negative, and no template controls) are run in duplicates for process control. Reference sequence for BRAF is NG_007873.3 RefSeqGene.
MLH1 Promoter Methylation Testing
At the North Shore Medical Center, MLH1 promoter methylation was performed at a CLIA approved commercial laboratory using a MethyLight assay,18 and a 10% value of the percentage of methylated reference was used as a cut-off. For samples from the Massachusetts General Hospital, MLH1 promoter methylation was performed using genomic DNA extracted from formalin-fixed paraffin-embedded tumor sections using methylation-specific PCR (MSP) at the CLIA-approved Laboratory of Molecular Diagnostics. Following bisulfite treatment (EZ DNA Methylation Kit, Zymo Research, Irvine, CA) of 500 ng to 1 μg of DNA, MSP is carried out with custom validated PCR primers specific to methylated and also to unmethylated MLH1 promoter sequences: MLH1 MSP ME F: 5′-GCGGTCCCAAAAGGGTCAGTCGGATAGCGATTTTTAACGC-3′; MLH1 MSP ME R: 5′-CCTAAAACGACTACTACCCG-3′; MLH1 MSP UN F: 5′-TAAAAATGAATTAATAGGAAGAGTGGATAGTG-3′; MLH1 MSP UN R: 5′-AATCTCTTCATCCCTCCCTAAAACA-3′. Forty-cycle PCR was performed as follows: denaturation 94°, 30 s; annealing 62°, 30 s; extension 72°, 1 min. The PCR products were analyzed using a 1.5% agarose gel.
Statistical Analysis
Continuous variables in patients’ baseline characteristics are presented as mean±standard deviation. The accuracy of BRAF mutation in predicting MLH1 promoter methylation was evaluated in the entire group and in subgroups stratified by tumor location, gender, and age. The analysis was performed to identify referral rates of three screening strategies: performing only BRAF mutation testing for all patients with absent MLH1 staining and referral for genetic evaluation and MLH1 sequencing for all BRAF− wild-type cases (strategy 1), performing only MLH1 promoter methylation testing for all patients with absent MLH1 stain with subsequent genetic evaluation for all negative cases (strategy 2), or using a stepwise two-tier approach, in which all cases with absent MLH1 are tested for a BRAF mutation, and MLH1 promoter methylation is performed only in BRAF− wild-type cases, and genetic counseling visit and testing is reserved for cases who tested negative for both (strategy 3).
Proportions, sensitivity, specificity, negative and positive predictive values are presented as percentages with 95% confidence interval) with continuity correction. Exact (Binomial) confidence interval was calculated for small-count groups. Comparison of age between study subgroups was performed using non-paired Student’s t-test, and comparison of dichotomous variables and proportions was done using Chi-square test, and Fisher exact probability test for small-count groups. A P-value of 0.05 or less was considered significant.
Results
Study Group
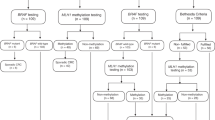
A total of 1011 consecutive colorectal cancers underwent immunohistochemical staining for all four mismatch repair proteins. Immunohistochemical staining identified 148 (14.6%, 95% confidence interval: 12.5–17%) cases with absent MLH1 staining. Most of these tumor samples failed to stain for both MLH1 and PMS2 (n=139). Four had weak MLH1 and absent PMS2 staining, and four others failed to stain for MLH1, PMS2, and MSH6. One sample exhibited loss of all four proteins. One hundred and twenty-six of these samples with abnormal MLH1 staining had both BRAF mutation and MLH1 promoter methylation testing completed successfully, and these were included in the final analysis (Figure 1).
Study flow, describing sub-grouping, tests’ results, and rates of genetic counseling, testing, and diagnosis. (+), positive; (−), negative; AMS, Amsterdam-II criteria; IHC, immunohistochemistry; Meth, promoter methylation; MMR, mismatch repair proteins; RBC, Revised Bethesda criteria; VUS, variant of unknown significance.
MLH1 Promoter Methylation and BRAF Mutation Testing
Among the 126 samples in which both MLH1 promoter methylation and BRAF testing were performed, three subgroups emerged. Eighty-six of the 126 samples (68.3%, 95% confidence interval: 59.3–76.1%) exhibited both a BRAF mutation and MLH1 promoter methylation, supporting the diagnosis of a sporadic tumor (group A). Sixteen samples (12.7%, 95% confidence interval: 7.7–20.1%) did not exhibit a BRAF mutation or MLH1 methylation, suggestive of a diagnosis of Lynch syndrome (group B). Twenty-four samples (19.1% 95% confidence interval:12.8–27.2%) exhibited discordant results (group C)—23 with MLH1 methylation but no BRAF mutation, and 1 with a BRAF mutation but no MLH1 promoter methylation.
The patients’ characteristics are detailed in Table 1. The age of diagnosis of cancer was similar in groups A and C, but patients in group B were significantly younger (P<0.001) and more likely to be male. All the four patients who met Amsterdam criteria were in group B. None of the patients in group C met Amsterdam or revised Bethesda criteria. Tumors were most commonly located in the right colon in all the three groups.
The correlation between BRAF genotype and MLH1 promoter methylation is illustrated in Table 2. Overall, the concordance between the two tests was 81% (95% confidence interval: 72.8–87.2%). The sensitivity of a BRAF mutation for MLH1 promoter methylation was 78.9% (95% confidence interval: 69.8–85.9%) with a specificity of 94.1% (95% confidence interval: 71.3–99.9%). The positive predictive value of a BRAF mutation for predicting MLH1 promoter methylation was 98.9% (95% confidence interval: 93.8–100%), but the negative predictive value was only 41% (95% confidence interval: 26–57.8%). When the subjects were segregated by age, the positive predictive value remained high at 92.3% (95% confidence interval: 64–99.8%) in patients under 70 years old, and the negative predictive value improved to 68.4% (95% confidence interval: 43.5–86.4%), though this did not reach statistical significance (P=0.09). However, the negative predictive value among patients ≥70 years old fell to 15% (95% confidence interval: 3.2–37.9%), which was significantly lower than that of younger patients (P<0.05). When tumors were separated by anatomic location in the colon, there was greater concordance in right-sided compared with left-sided tumors (82.8% (95% confidence interval: 73.3%–89.6%) vs 72.7% (95% confidence interval: 39%–94%), respectively). However, there was no significant change in the positive and negative predictive values.
When patients were stratified according to gender, the negative predictive value of a BRAF mutation in predicting MLH1 promoter methylation was greater in males (67% (95% confidence interval: 38.7–87%) compared with females (25% (95% confidence interval: 10.6–47.1%), P<0.05). This was particularly apparent in older female patients. In women >70 years who did not exhibit a BRAF mutation in their tumor, nearly all were positive for MLH1 promoter methylation, resulting in a negative predictive value of only 6.7% (95% confidence interval: 0.2–31.9%; Table 2).
Results of Genetic Testing
Formal genetic evaluation for Lynch syndrome was recommended for the 40 individuals in groups B (n=16) and C (n=24). This represents about one-third (31.7% (95% confidence interval: 23.9–40.7%)) of the 126 patients who had complete BRAF and MLH1 methylation testing. In group B, six of the cases met the revised Bethesda criteria and four of the cases were positive for Amsterdam II criteria. No cases from group C met either revised Bethesda or Amsterdam I or II criteria.
In group B, 13 individuals followed through with a genetic counseling visit, and germline genetic testing was performed in 10 patients. One pathogenic MLH1 mutation was identified in a 52-year-old female with an ascending colon tumor who met the Amsterdam criteria. In group C, nine individuals attended a genetic counseling session, and six pursued genetic testing. In this group, no pathogenic mutations were identified, and only a variant of unknown significance in MLH1 (ref. 19) was identified in a 50-year-old whose tumor was BRAF wild-type/MLH1 methylation positive. This patient did not have any first- or second-degree relatives with either colorectal or endometrial cancer. No pathogenic MLH1 mutations were identified in any individual from group C.
Of note, two patients with absence of MLH1 protein on initial immunohistochemistry were members of established Lynch syndrome families. BRAF and MLH1 promoter methylation testing were deferred, and the patients were directly referred for genetic testing. Hence, they were not included in group A, B, or C. Both individuals were subsequently found to carry the respective familial MLH1 gene mutation.
Modeling a Two-Step Approach to Universal Screening
We considered several management strategies for the evaluation of loss of MLH1 protein on immunohistochemistry. Three approaches were evaluated with respect to anticipated rates of referral for genetic counseling: (strategy 1) MLH1 promoter methylation testing alone, (strategy 2) BRAF testing alone, or (strategy 3) BRAF testing followed by MLH1 promoter methylation testing for BRAF wild-type cases. In our study group, 86.5% of 126 patients (95% confidence interval: 79–91.7%) with abnormal MLH1 staining tested positive for MLH1 promoter methylation. This strategy 1 would translate to a referral rate of 13.5% (95% confidence interval: 8.3–21%) for further genetic counseling and testing. With respect to BRAF testing (strategy 2), 69% (95% confidence interval: 60.1–76.8%) of cases with loss of MLH1 immunostaining tested positive for a BRAF mutation, and the remaining 31% (95% confidence interval: 23.2–39.9%) would have required subsequent genetic counseling and testing (Figure 2). Utilization of BRAF as a sole test would have significantly increased the number of patients referred to genetic counseling by 2.3-fold (P<0. 01).
A schematic representation of the proportion of patients that would have been referred to genetic counseling for suspected LS using a (a) single test approach utilizing either MLH-1 methylation or BRAF mutation testing, or a (b) hybrid approach in which BRAF mutation negative patients are tested for MLH1 methylation.
MLH1 promoter methylation testing is more expensive and less accessible than BRAF genotyping. We, therefore, considered a two-step approach in which BRAF testing is first offered to all patients with abnormal MLH1 staining, and MLH1 promoter methylation testing is reserved for those who test negative for a BRAF mutation. Further genetic testing is recommended only for those who then test negative for MLH1 promoter methylation. In this strategy 3, the rate of patients referred to genetic counseling was significantly lower than that with the BRAF testing-only strategy (P<0.001), and was similar to the MLH1 promoter methylation-only strategy. In addition, there was a 69% reduction in the number of methylation tests performed.
Discussion
In a large, prospective universal Lynch syndrome screening program, loss of MLH1 staining was the most common finding. We sought to define the performance characteristics of BRAF and MLH1 promoter methylation testing in this setting. Our results indicate that there is an 81% concordance between BRAF genotype and MLH1 promoter methylation. In addition, there was a high positive predictive value for BRAF mutation predicting MLH1 promoter methylation and a low negative predictive value, primarily in female patients >70 years old. A hybrid strategy is proposed in which BRAF mutation analysis is first performed and cases without a BRAF mutation then undergo MLH1 promoter methylation analysis rather than direct referral for genetic counseling.
Lynch syndrome is under-recognized in the population, and screening for the condition can ultimately reduce the risks for cancer in a cost-effective manner.20 BRAF genotyping is currently the most commonly used test in the evaluation of colon cancer cases with loss of MLH1 staining. Our findings suggest that such an approach increases the number of referrals of low-risk patients for Lynch syndrome genetic testing over 2-fold. The costs associated with genetic counseling and testing are significant. Testing for MLH1 promoter methylation is redundant in cases that test positive for a BRAF mutation, and a hybrid approach in which MLH1 promoter methylation is reserved for BRAF wild-type cases would lead to a lower referral rate for genetic counseling compared with BRAF testing only and to fewer methylation tests performed compared with a strategy of methylation testing only. Refining strategies to minimize the number of screening tests performed as well as referrals of low-risk patients for genetic testing can reduce these costs and thereby serve to improve the performance and uptake of such a screening program.
Nearly 20% of cases exhibited a discordant pattern between BRAF and MLH1 promoter methylation results. This is similar to a prior report by Newton and colleagues that revealed a 17.4% discordance rate (25 of 144) in selected colorectal cancer cases with loss of MLH1 on immunohistochemistry.14 Our study included consecutive, non-selected patients, and none of these patients with discordant BRAF/MLH1 methylation results were ultimately given a diagnosis of Lynch syndrome by either genetic or clinical criteria. Although not all had blood genetic testing performed, their clinical features and family history were not consistent with a diagnosis of Lynch syndrome. In the previous report, the vast majority of patients with a germline MLH1 mutation who had discordant tumor testing results did indeed meet Amsterdam criteria.14 The average age of cancer diagnosis in this group was 75.4 years and none met Amsterdam or Bethesda criteria. There were two cases in which there was a family history of colon or endometrial cancer in a single first-degree relative. Germline testing (for mutations in MLH1 and PMS2) was performed in both these cases, and no mutations were identified.
The vast majority of these discordant cases exhibited MLH1 promoter methylation with wild-type BRAF, consistent with prior reports,14 suggesting that MLH1 promoter methylation may serve as a more accurate marker for a sporadic colon tumor, particularly in individuals over 70 years of age. It should be noted, however, that methylation of the MLH1 gene promoter has been reported in a small subset of tumors from individuals with Lynch syndrome.14, 21 This may reflect somatic inactivation of the second MLH1 allele or in very rare cases, germline MLH1 promoter methylation. These reports were derived from highly selected populations and in our large screening population, a diagnosis of Lynch syndrome was not confirmed in any of these cases with MLH1 promoter methylation. It is also important to emphasize that molecular screening algorithms should not supersede clinical impressions generated by a strong family history or very young age of cancer diagnosis, and genetic counseling should be recommended if such clinical suspicions are present.
The pathophysiological mechanisms that determine whether a tumor with MLH1 promoter methylation acquires a BRAF mutation or not are undefined. Although both groups have a favorable natural history when compared with microsatellite-stable (MSS) tumors, there are distinctive tumor behaviors between these two molecular subtypes, and a more favorable prognosis has been described in BRAF wild-type tumors with microsatellite instability.22
In our population, the overall germline mutation rate was low, even in group B, limiting a complete discussion about the correlations between MLH1 promoter methylation, BRAF mutations, and germline MLH1 mutations. The uptake of genetic testing was suboptimal in group B (62.5%) but was higher than prior reports, in which a 26% rate was observed when using a similar strategy.23 In addition, these data indicate the rate of germline MLH1 mutations only and therefore do not reflect the overall rate of Lynch syndrome owing to mutations in MSH2, MSH6, PMS2, or EPCAM.
In conclusion, there is an inconsistent correlation between BRAF and MLH1 promoter methylation tests in a Lynch syndrome screening program in individuals over 70 years of age with diagnosis of colon cancer. A hybrid approach in which MLH1 promoter methylation is reserved for BRAF wild-type cases could reduce the number of low-risk patients referred to genetic counseling. Such an approach may reduce costs associated with universal screening for Lynch syndrome and facilitate its widespread implementation. Regardless of the approach, attention to clinical features that include age of cancer diagnosis and family history should remain part of the overall evaluation.
References
Group USCSW. United States Cancer Statistics: 1999–2012 Incidence and Mortality Web-based Report. Atlanta: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention and National Cancer Institute. 2015, Available at: www.cdc.gov/uscs. In: Vol. 2016.
Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J et al, Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 2013;49:1374–1403.
Haggar FA, Boushey RP . Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg 2009;22:191–197.
Giardiello FM, Allen JI, Axilbund JE et al, Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol 2014;109:1159–1179.
Vasen HF, Blanco I, Aktan-Collan K et al, Revised guidelines for the clinical management of Lynch syndrome (HNPCC): recommendations by a group of European experts. Gut 2013;62:812–823.
Evaluation of Genomic Applications in Practice and Prevention Working G. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 2009;11:35–41.
Zhang X, Li J . Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol 2013;5:12–19.
Kidambi TD, Blanco A, Myers M et al, Selective versus universal screening for lynch syndrome: a six-year clinical experience. Dig Dis Sci 2015;60:2463–2469.
Boland CR, Goel A . Microsatellite instability in colorectal cancer. Gastroenterology 2010;138:2073–2087.e3.
Toon CW, Walsh MD, Chou A et al, BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol 2013;37:1592–1602.
Weissman SM, Burt R, Church J et al, Identification of individuals at risk for Lynch syndrome using targeted evaluations and genetic testing: National Society of Genetic Counselors and the Collaborative Group of the Americas on Inherited Colorectal Cancer joint practice guideline. J Genet Couns 2012;21:484–493.
Jin M, Hampel H, Zhou X et al, BRAF V600E mutation analysis simplifies the testing algorithm for Lynch syndrome. Am J Clin Pathol 2013;140:177–183.
Parsons MT, Buchanan DD, Thompson B et al, Correlation of tumour BRAF mutations and MLH1 methylation with germline mismatch repair (MMR) gene mutation status: a literature review assessing utility of tumour features for MMR variant classification. J Med Genet 2012;49:151–157.
Newton K, Jorgensen NM, Wallace AJ et al, Tumour MLH1 promoter region methylation testing is an effective prescreen for Lynch Syndrome (HNPCC). J Med Genet 2014;51:789–796.
Syngal S, Brand RE, Church JM et al, ACG clinical guideline: genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 2015;110:223–262 quiz 63.
Mangold E, Pagenstecher C, Friedl W et al, Tumours from MSH2 mutation carriers show loss of MSH2 expression but many tumours from MLH1 mutation carriers exhibit weak positive MLH1 staining. J Pathol 2005;207:385–395.
Dias-Santagata D, Akhavanfard S, David SS et al, Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med 2010;2:146–158.
Ogino S, Kawasaki T, Brahmandam M et al, Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. The Journal of molecular diagnostics. J Mol Diagn 2006;8:209–217.
Sijmons RH, Greenblatt MS, Genuardi M . Gene variants of unknown clinical significance in Lynch syndrome. An introduction for clinicians. Fam Cancer 2013;12:181–187.
Mvundura M, Grosse SD, Hampel H et al, The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med 2010;12:93–104.
Moreira L, Munoz J, Cuatrecasas M et al, Prevalence of somatic mutl homolog 1 promoter hypermethylation in Lynch syndrome colorectal cancer. Cancer 2015;121:1395–1404.
Lochhead P, Kuchiba A, Imamura Y et al, Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst 2013;105:1151–1156.
Heald B, Plesec T, Liu X et al, Implementation of universal microsatellite instability and immunohistochemistry screening for diagnosing lynch syndrome in a large academic medical center. J Clin Oncol 2013;31:1336–1340.
Acknowledgements
The authors would like to acknowledge Dr Hui Zheng of the the Massachusetts General Hospital Biostatistics Center and Harvard Catalyst (The Harvard Clinical and Translational Science Center) for his statistical support. TA is a recipient of a fellowship grant from The American Physicians Fellowship for Medicine in Israel and from Israel Cancer Association.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr Lauwers has received consulting fees from Biogen, Inspirata, and Nine Point. Dr Iafrate has received royalties from ArcherDx.
Rights and permissions
About this article
Cite this article
Adar, T., Rodgers, L., Shannon, K. et al. A tailored approach to BRAF and MLH1 methylation testing in a universal screening program for Lynch syndrome. Mod Pathol 30, 440–447 (2017). https://doi.org/10.1038/modpathol.2016.211
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2016.211
This article is cited by
-
PREMM5 distinguishes sporadic from Lynch syndrome-associated MMR-deficient/MSI-high colorectal cancer
Familial Cancer (2023)
-
Evaluating mismatch repair deficiency in colorectal cancer biopsy specimens
Histochemistry and Cell Biology (2023)
-
Comparison of universal screening in major lynch-associated tumors: a systematic review of literature
Familial Cancer (2022)
-
Worldwide variation in lynch syndrome screening: case for universal screening in low colorectal cancer prevalence areas
Familial Cancer (2021)
-
Comparisons of screening strategies for identifying Lynch syndrome among patients with MLH1-deficient colorectal cancer
European Journal of Human Genetics (2020)