Abstract
When distinguishing between indolent and potentially harmful prostate cancers, the Gleason score is the most important variable, but may be inaccurate in biopsies due to tumor under-sampling. This study investigated whether a molecular feature, PTEN protein loss, could help identify which Gleason score 6 tumors on biopsy are likely to be upgraded at radical prostatectomy. Seventy one patients with Gleason score 6 tumors on biopsy upgraded to Gleason score 7 or higher at prostatectomy (cases) were compared with 103 patients with Gleason score 6 on both biopsy and prostatectomy (controls). A validated immunohistochemical assay for PTEN was performed, followed by fluorescence in situ hybridization (FISH) to detect PTEN gene deletion in a subset. PTEN protein loss and clinical–pathologic variables were assessed by logistic regression. Upgraded patients were older than controls (61.8 vs 59.3 years), had higher pre-operative PSA levels (6.5 vs 5.3 ng/ml) and a higher fraction of involved cores (0.42 vs 0.36). PTEN loss by immunohistochemistry was found in 18% (13/71) of upgraded cases compared with 7% (7/103) of controls (P=0.02). Comparison between PTEN immunohistochemistry and PTEN FISH showed the assays were highly concordant, with 97% (65/67) of evaluated biopsies with intact PTEN protein lacking PTEN gene deletion, and 81% (13/16) of the biopsies with PTEN protein loss showing homozygous PTEN gene deletion. Tumors with PTEN protein loss were more likely to be upgraded at radical prostatectomy than those without loss, even after adjusting for age, preoperative PSA, clinical stage and race (odds ratio=3.04 (1.08–8.55; P=0.035)). PTEN loss in Gleason score 6 biopsies identifies a subset of prostate tumors at increased risk of upgrading at radical prostatectomy. These data provide evidence that a genetic event can improve Gleason score accuracy and highlight a path toward the clinical use of molecular markers to augment pathologic grading.
Similar content being viewed by others
Main
Based on pathologic examination of the entire prostate, the Gleason grading system is the most powerful variable available for distinguishing between indolent Gleason score 3+3=6 cancers and cancers with lethal potential, virtually all of which are Gleason score 3+4=7 or higher.1, 2, 3, 4, 5 If the Gleason score of the needle biopsy accurately reflected that of the dominant tumor, it would be relatively simple to identify appropriate candidates for deferred treatment or active surveillance in place of definitive therapy. However, a recent meta-analysis, including >14 000 cases, suggests that this is frequently not the case: a mean of 36% (range: 14–51%) of apparently indolent Gleason score 6 tumors on needle biopsy are ‘upgraded’ to Gleason score 7 or higher at radical prostatectomy.6
Currently, there are few predictors of which patients with Gleason score 6 prostate tumors on biopsy are likely to harbor higher-grade disease. Tumor upgrading from biopsy to radical prostatectomy can occur for multiple reasons, including tumor progression, pathology errors, and inter-observer variability in Gleason grading. However, a common cause is under-sampling of the highest grade cancer.6 Unfortunately, increasing the number of biopsy cores above the current standard of 12 does not substantially reduce the frequency of upgrading.7 While multiparametric MRI-based imaging is becoming more promising in this regard,8 MRI imaging is expensive and not widely used. Further, prior studies have found that clinical–pathologic parameters such as higher preoperative PSA, lower prostate volume, and more extensive disease on biopsy are relatively weak predictors of upgrading.9, 10, 11, 12, 13, 14, 15 More importantly, to date, no molecular biomarkers have been shown to correlate with upgrading from Gleason score 6 to Gleason score 7.
We and others have shown that expression of PTEN, a tumor suppressor, is most commonly lost in prostate tumors with high Gleason grade and stage, frequently as a result of PTEN gene deletion.16, 17, 18, 19, 20, 21, 22, 23, 24, 25 Recently, we developed and validated a simple and inexpensive immunohistochemical assay to assess PTEN protein status in prostate cancer.16 Using this assay, we showed that PTEN protein loss, assessed by a simple dichotomous scoring system, is highly correlated with underlying genetic deletions of the PTEN gene and is associated with increased Gleason grade and decreased time to biochemical recurrence, disease progression and metastasis in contemporary surgical cohorts.16, 17, 18 Here, we demonstrate that PTEN protein loss in Gleason score 6 needle biopsy specimens, although uncommon, is associated with increased risk of Gleason score upgrading in the final radical prostatectomy specimen.
Materials and methods
Patients and Tissue Samples
With institutional review board approval, pathology databases were queried for all needle biopsies performed from 2000 to 2011 at Johns Hopkins Hospital, a tertiary care hospital, and Johns Hopkins Bayview Medical Center, a community hospital, that contained only Gleason score 6 followed by a radical prostatectomy specimen that contained Gleason score 7 or higher. It is important to note that at Johns Hopkins Hospital, <25% of radical prostatectomy patients had biopsies also performed at Johns Hopkins Hospital, which meant that 75% of radical prostatectomy cases at Johns Hopkins Hospital did not have biopsy tissue available for study. In contrast, at Bayview Medical Center, the proportion with available biopsies was >60%. Controls with Gleason score 6 on biopsy followed by Gleason score 6 tumor on radical prostatectomy were selected as sequential biopsies occurring between 2000 and 2004 at Johns Hopkins Hospitals and between 2000 and 2011 at Bayview Medical Center (Supplementary Figure 1). As is routine, all radical prostatectomy tissue was entirely submitted for histologic analysis. All biopsies and radical prostatectomy slides were re-reviewed and re-graded by trained uropathologists (DMB and TLL) using the 2005 modified International Society of Urologic Pathology (ISUP) grading system.26 It is not universally agreed upon whether a tumor with majority Gleason pattern 3 and <5% Gleason pattern 4 should be graded as Gleason 3+3=6 with tertiary pattern 4 or Gleason 3+4=7. In favor of using a tertiary grade, the ISUP consensus paper noted that tumors with majority Gleason pattern 3 and <5% pattern 4 generally have a lower pathologic stage than a Gleason score 3+4=7 where pattern 4 occupies >5% of the tumor.26 Overall, in the current study, 40% (41/103) of the non-upgraded control radical prostatectomies had a component (<5%) of tertiary Gleason pattern 4. However, because the inter-observer reproducibility of tertiary grading remains largely untested and because Gleason score 6 tumors have negligible lethality regardless of tertiary Gleason pattern,1 grading in the current study was defined solely by Gleason score without reference to a tertiary component. A single block (usually comprising two cores) containing the largest percentage involvement by tumor was selected for PTEN immunostaining.
PTEN Immunohistochemistry and Interpretation
PTEN immunohistochemistry was performed manually as previously described and blindly scored by two uropathologists (TLL and DMB) using a validated dichotomous scoring system.16 In brief, 4-μm biopsy sections were deparaffinized and re-hydrated under standard conditions. Antigen unmasking was performed by steaming in EDTA buffer (pH 8.0) for 45 min. Endogenous peroxidase activity was quenched by incubation with peroxidase block for 5 min at room temperature. Slides were incubated for 45 min at room temperature with a rabbit anti-human PTEN antibody (Clone D4.3 XP; Cell Signaling, Danvers, MA, USA; 1:50 dilution). A horseradish peroxidase-labeled polymer (PowerVision, Leica Microsystems, Bannockburn, IL, USA) was applied for 30 min at RT and signal detection was performed using 3,3'-diaminobenzidine tetrahydrochloride (DAB) as the chromagen. Slides were counterstained with hematoxylin, dehydrated, and mounted.
PTEN immunohistochemistry on each case was independently scored by two uropathologists (TLL and DMB) using a previously validated dichotomous scoring system.16 Cases were considered to have PTEN protein loss if the intensity of cytoplasmic and nuclear staining was markedly decreased or entirely negative across >10% of tumor cells compared with surrounding benign glands and/or stroma. As we have seen previously, some cases with markedly decreased cytoplasmic staining showed intact staining on the apical membrane of the cell (Figures 1b, d and 2F). The biologic significance of this staining in the context of homozygous deletions of the PTEN gene is unclear (Figure 2). While we cannot entirely exclude issues of antibody specificity, immunoblotting of human prostate lysates with this antibody identifies a dominant band at the expected molecular weight, suggesting the antibody is quite specific for PTEN protein in this tissue. Additionally, it remains unclear whether this apical staining represents intra- or extracellular protein. Importantly, despite this apical staining, we have shown that the inter-observer reproducibility of the dichotomous scoring system developed for cytoplasmic/nuclear PTEN protein remains high16 with disagreements about the immunohistochemistry scoring in only 17/174 or 10% of cases in the current study. In cases of disagreement, a third uropathologist (AMD) blindly scored the case to break the tie.
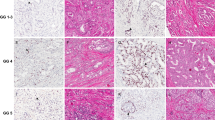
Gleason Score 6 prostate biopsies that are upgraded to Gleason Score 7 at radical prostatectomy show PTEN protein loss by immunohistochemistry (a) Low power ( × 200 magnification) photomicrograph of PTEN immunohistochemistry in a Gleason score 6 needle biopsy specimen demonstrates PTEN protein loss in tumor glands (arrow) with preservation of PTEN staining in intermingled benign glands (arrowheads). (b) High power ( × 630) image highlights infiltrating tumor glands with PTEN loss and surrounding benign glands with intact staining. Note the presence of apical membrane staining in glands with cytoplasmic PTEN loss (arrowhead). The biological significance of this membrane staining remains unclear. (c) Low power ( × 200 magnification) photomicrograph of tumor with heterogeneous PTEN protein loss, demonstrating PTEN protein loss in some tumor glands (arrow) and intact PTEN protein in adjacent tumor (arrowhead). (d) High power ( × 630) image showing tumor glands with and without PTEN protein loss. As in b, note the presence of apical membrane staining in glands with cytoplasmic PTEN loss (arrowhead). The biological significance of this membrane staining remains unclear.
PTEN FISH results are highly concordant with PTEN immunohistochemistry (a) PTEN immunohistochemistry image ( × 200 magnification) demonstrating PTEN protein loss in tumor glands (arrow) and PTEN protein retention in nearby benign glands (arrowhead) (b) PTEN FISH image captured from the tumor in (a) demonstrates glands with homozygous PTEN gene loss (yellow signal; arrow and upper right panel) intermixed with glands with PTEN intact (yellow signal; arrowhead, lower right panel). Peri-centromeric control probes (red) as well as flanking gene probes WAPAL (green) and FAS (aqua) are intact in all cells. (c) PTEN immunohistochemistry image ( × 200 magnification) from a separate Gleason score 6 biopsy that was not upgraded at radical prostatectomy demonstrating focal loss of cytoplasmic PTEN protein in tumor glands (arrow). Adjacent tumor glands stain positively. (d) PTEN FISH image captured from tumor in (c) demonstrates lack of PTEN deletion in tumor cells throughout (inset). (e) PTEN immunohistochemistry image ( × 100) demonstrates PTEN protein loss in tumor glands (red outline) with PTEN protein retention in adjacent benign glands (blue outline). (f) Higher power ( × 630) image of boxed area from (e). Note the presence of apical membrane staining in glands with cytoplasmic PTEN loss (arrowhead). The biological significance of this membrane staining remains unclear in the context of homozygous deletion of the PTEN gene. (g) PTEN FISH image captured from region delineated in (e) demonstrates intermixed glands with hemizygous PTEN, WAPAL and FAS deletion (arrow, inset) and glands lacking PTEN deletion (arrowhead).
PTEN FISH
Four-color FISH was performed and interpreted as described previously using the PTEN del-TECT probe (CymoGen Dx, New Windsor, NY, USA; ref. 23; www.ptendeletion.net). An H&E-stained needle core section was available for comparison to confirm regions of interest. In cases where PTEN deletions were identified, >60% of nuclei within a given focus exhibited the aberration.
Statistical Analysis
Means and proportions of pre- and post-operative characteristics of cases and controls were compared using the two-sample t-test and the χ2 test, respectively. Multivariable logistic regression was used to estimate the odds ratios and 95% confidence intervals of a biopsy upgrade among men with PTEN protein loss. First, odds ratios were estimated adjusted for age at diagnosis. Next, results were adjusted for preoperative PSA (continuous, log-transformed) and clinical stage (binary, T2 or higher). Results were further adjusted for race (binary, nonwhite). Statistical analyses were performed using SAS 9.3 (Cary, NC, USA).
Results
Pre-Operative Clinical–Pathologic Parameters
The mean age of the 71 upgraded cases was slightly older than that of the 103 controls who were not upgraded (61.8 vs 59.3 years; P=0.005; Table 1). The racial makeup of the two groups was not significantly different. The mean pre-operative serum PSA level was 23% higher in the upgraded cases (6.5 vs 5.3 ng/ml; P=0.009), but there was no difference in clinical stage distribution between the two groups. The majority of patients underwent radical prostatectomy within 3 months of biopsy diagnosis and there was no significant difference in the pre-operative interval between groups.
Greater than 90% of both cases and controls had a 12-core biopsy performed, with a comparable number of cores examined in the cases compared to the controls (12.2 vs 11.9; P=0.06). The fraction of cores involved by tumor was slightly higher in the cases (0.36 vs 0.30; P=0.04); however, the maximum percent tumor per core was not significantly different between groups. Tumor was present on bilateral cores (from the left and right prostate) in a higher percentage of the upgraded cases compared with controls (55 vs 41%, P=0.07). Finally, the fraction of cases with perineural invasion was not significantly different between groups.
Post-Operative Clinical–Pathologic Parameters
To validate the clinical significance of tumor upgrading, post-operative clinical–pathologic parameters for the two groups of patients were assessed (Table 2). Upgraded cases had smaller mean prostate weights at radical prostatectomy (54 vs 60 g), although this did not reach statistical significance (P=0.17). Location of the dominant tumor nodule was assessed in 94% (67/71) of upgraded patients and was divided between anterior-predominant nodules (33% or 22/67) and posterior/lateral-predominant nodules (67% or 45/67). The proportion of anterior-dominant tumors in the upgraded Gleason score 6 cases was similar to that seen in other unselected biopsy Gleason score 6 populations at Johns Hopkins27, 28 and elsewhere, suggesting a lack of significant enrichment for anterior-dominant tumors in this sample.29 As expected, upgraded cases were significantly more likely to have had extraprostatic extension or seminal vesicle involvement at radical prostatectomy than controls (35 vs 18%; P=0.01). Accordingly, upgraded cases were more likely to have positive margins at radical prostatectomy (24 vs 8%; P=0.003) as well as PSA recurrence (9 vs 1%; P=0.03), with a mean follow-up period of 3.6 and 4.5 years respectively for the ∼80% of patients with available information in each group.
PTEN Protein Loss and PTEN Gene Status
PTEN protein loss was observed in 18.3% (13/71) of the upgraded tumors compared with only 7% (7/103) of the controls (P=0.02). A subgroup analysis of the two hospital populations included in the study showed nearly identical results in both patient populations (18 and 20% for cases in Johns Hopkins Hospitals and Bayview Medical Center, respectively, and 7 and 6% for controls, respectively). PTEN protein loss was frequently focal within a given tumor sample (Figures 1 and 2) with heterogeneous PTEN staining in 85% (11/13) of the upgraded cases with PTEN protein loss and 100% (7/7) of the controls.
A recently developed four-color FISH assay23 was used to assess for PTEN gene deletions in the 20 biopsies with decreased PTEN protein by immunohistochemistry. PTEN FISH was evaluable in 86% of the non-upgraded controls (6/7) and 77% (10/13) of the upgraded cases. Biopsies that could not be evaluated included those where the FISH hybridization failed (n=2) or where the tumor area with decreased PTEN protein could not be aligned on the adjacent histologic section on which the FISH was performed (n=2). Among the evaluable upgraded cases, 90% (9/10) of the biopsies with decreased PTEN protein showed homozygous PTEN deletions by FISH, variably involving the adjacent gene probes for FAS and WAPAL (Figure 2). Only one upgraded case (10%) with PTEN protein loss lacked PTEN gene deletion as assessed by FISH. Among the evaluable non-upgraded controls that showed unambiguous loss of PTEN protein by immunohistochemistry, 67% had homozygous PTEN gene deletions (4/6), while 17% (1/6) had a hemizygous deletion and 17% (1/6) had no apparent PTEN deletion. Of the cases with PTEN gene deletion, this deletion was focal (involving only a subset of tumor glands) in 87% (13/15) cases, consistent with focal loss of immunohistochemical staining in a similar proportion of cases (90% or 18/20). In cases where it was possible to directly match individual tumor glands on the two different slides separated by 4 μm, focal PTEN loss by immunohistochemistry was concordant with focal gene loss by FISH (Figure 2).
We also performed PTEN FISH in a subset of 76 biopsies with intact PTEN protein. These biopsies were randomly selected for FISH and represented 51% (49/96) of the control biopsies with intact PTEN protein and 47% (27/58) of the upgraded cases with intact PTEN protein. PTEN FISH was evaluable in 92% (45/49) of the controls and 81% (22/27) of the upgraded cases. Biopsies that could not be evaluated included those where the FISH hybridization failed (n=8) and one case where there was no tumor remaining for evaluation on deeper sections. Of 67 biopsies with intact PTEN protein and evaluable PTEN FISH, 97% (65/67) lacked deletion of the PTEN gene, while two biopsies (3%) had hemizygous deletion of PTEN, both of which were upgraded cases. Thus, we conclude that in the representative subset studied, PTEN immunohistochemistry and PTEN FISH results were highly concordant. While 97% (65/67) of biopsies with intact PTEN protein lacked PTEN gene deletion (and none showed homozygous gene deletion), 81% (13/16) of the biopsies with PTEN protein loss showed homozygous PTEN gene deletion.
Association of PTEN Protein Loss and Upgrading
Logistic regression was performed to assess whether PTEN protein loss was independently associated with Gleason score upgrading (Table 3). The variables adjusted in the multivariable analysis were those that have been found to be associated with upgrading in the current and multiple prior studies, including patient age and race, pre-operative PSA level and clinical stage. Even after adjusting for all of these variables (Model B), the odds ratio of Gleason score upgrading was 3.04 (95% confidence interval: 1.08–8.55; P=0.035) for PTEN protein loss.
Discussion
The Gleason grading system remains one of the most powerful prognostic indicators in prostate cancer, yet tumor under-sampling remains an important limitation of the Gleason system applied to biopsy specimens. Here, we have shown that PTEN protein loss in Gleason score 6 biopsies, although uncommon, is associated with an increased risk of tumor upgrading at radical prostatectomy. The association of PTEN protein loss with tumor upgrading suggests that, at least in a subset of patients, Gleason pattern 3 tumor associated with adjacent pattern 4 differs at the genomic level from pattern 3 derived from a purely Gleason score 6 tumor.24, 30 Indeed, using radical prostatectomy specimens, we have found that PTEN protein loss in pattern 3 carcinoma from a Gleason score 7 cancer is significantly more common than that of pattern 3 carcinoma from a purely Gleason score 6 tumor (AMD and BJ Trock, in preparation). These studies suggest that an integrated approach exploiting genomic markers in limited tumor biopsy specimens may be helpful to improve on the prognostic accuracy of our current pathologic grading system. Such improvements are sorely needed to avoid overtreatment of indolent prostate cancers.
Importantly, PTEN loss by immunohistochemistry is only expected to be present in ∼11% of Gleason score 6 biopsies overall based on the current study (using the upgrading rate of 25% from our institution6) and this is highly consistent with our prior studies of radical prostatectomy specimens.16 Thus, while PTEN protein loss (a ‘positive test’) suggests an increased risk of upgrading and potentially can be used to inform a treatment decision, the lack of loss (a ‘negative’ test) is likely to be much less informative as a result of inadequate tumor sampling and tumor heterogeneity. While we could not calculate the positive predictive and negative predictive values in this case–control design, it is likely that additional studies would show that the positive predictive value of PTEN protein loss for predicting upgrading will be significant enough to be useful, but that the negative predictive value of no loss in PTEN may not be as useful. Thus, for a molecular test to be clinically useful for both positive and negative predictive values, it is likely that a larger panel of molecular markers will be needed. Nevertheless, PTEN protein loss portends not only an increased risk of upgrading, but an increased risk of biochemical recurrence (independent of grade) following radical prostatectomy,17 as well as potential sensitivity to several classes of targeted therapies for phosphatidylinositol-3-kinase (PI3K) signaling. Thus, despite the relatively low expected prevalence of PTEN protein loss in the Gleason score 6 population, we anticipate that this relatively inexpensive test may become useful for one or more clinical indications.
In the current study, we report a very high correlation between PTEN immunohistochemistry and FISH results, with 87% of cases with PTEN protein loss showing underlying PTEN deletions and 97% of cases with intact protein lacking underlying deletions. This suggests that, at least in the current study population, loss of the PTEN gene is the major mechanism underlying loss of PTEN protein expression. Protein loss through mechanisms such as point mutation or epigenetic silencing would appear to be relatively rare, as shown recently.31, 32, 33 However rare, it is possible that some cancers lose PTEN activity with hemizygous loss of one allele and mutations in the remaining allele that yield a stable but functionally compromised protein.
PTEN deletions may take place earlier in prostate cancer oncogenesis than previously suspected.34, 35 We found focal loss in 14% of cases. Focal loss suggests that methods that rely on nucleic acid extraction might not be as sensitive in detecting PTEN loss as the in situ methods employed in this study. Moreover, recent refinements of the FISH technique have improved the sensitivity for detecting deletions, by incorporating closely linked flanking markers either side of the PTEN gene23 (www.ptendeletion.net). Overall, the rate of PTEN deletion using this probe design in our Gleason score 6 biopsy population is comparable to the rate reported in other recent biopsy cohorts assessed by FISH, where deletions (hemizygous and homozygous) are present in ∼10% of cases.36 However, given the general prevalence of hemizygous over homozygous deletions in previous PTEN FISH studies in localized prostate cancer,21, 24, 34 a surprising finding in the current study was that the majority of Gleason score 6 cases with PTEN protein loss showed homozygous PTEN gene deletion. Perhaps in part because of this, the PTEN immunohistochemistry assay appears to be highly concordant with underlying PTEN gene status. It remains to be seen whether this protein assay is similarly sensitive for detecting PTEN deletions in cohorts with higher rates of hemizygous PTEN gene loss. Indeed, immunohistochemistry may be relatively insensitive for detecting cases with subtly reduced levels of PTEN protein, because we only score cases as decreased if there is either total loss or the signal is ‘markedly decreased’ as compared with normal acini. Future studies will examine whether hemizygous deletions may predict upgrading. If so, performance of both immunohistochemistry and FISH may represent the optimal strategy for risk assessment.
Our study has a number of important strengths. Although PTEN immunohistochemistry was conducted manually in the current study, this protocol has been successfully replicated and validated on the Ventana Benchmark platform in a CLIA-certified laboratory (TLL and AMD, data not shown). Also relevant to clinical practice, 85% of patients in the current study would have met the extended criteria for active surveillance recently evaluated at Johns Hopkins.37 Previous studies have demonstrated that biopsies consisting of fewer than 10 cores are associated with significantly higher upgrading rates.12 The vast majority of patients (>90%) in our study underwent 12 core biopsy, which is currently the standard of care in the United States. Additionally, in our study, all radical prostatectomy tissue from each case was entirely submitted for histologic examination, ensuring accurate Gleason score assignment. Finally, all biopsies and radical prostatectomies were re-graded by urologic pathologists using the most updated (2005) Gleason scoring system.26 Our findings are further validated by the clinical–pathologic variables we found associated with upgrading, which are highly consistent with contemporary studies in larger cohorts, including patient age, pre-operative PSA, clinical stage and extent of tumor involvement on biopsy.9, 10, 11, 12, 13, 14, 15 As expected, upgraded patients in the current study also fared significantly worse after surgery, with substantially higher rates of extraprostatic extension, surgical margin positivity and biochemical recurrence compared to controls who were not upgraded at radical prostatectomy.
There are a few notable limitations of the current study, perhaps the most important of which is the relatively small sample size. Because of this, it is essential that our results be validated in future studies with larger, prospective cohorts. Partly mitigating this weakness, our study did include patients from two clinical settings (tertiary vs community care) yet each showed nearly identical rates of PTEN protein loss in upgraded cases and controls, suggesting that our findings will likely be reproducible in independent cohort analyses. In addition, because of the case–control design of the current study, it was not possible to formally evaluate the sensitivity and specificity of PTEN for predicting upgrading and to estimate what additional information PTEN adds above and beyond currently collected clinical–pathologic variables. However, our multivariable analysis suggests that PTEN remains significantly associated with upgrading even when controlling for clinical stage, race and PSA. An additional potential weakness is that we evaluated only one to two needle biopsy cores for most patients. As we and others have showed that PTEN protein loss and underlying gene deletion is most commonly focal,24, 35 it is likely that we missed a number of cases with focal PTEN protein loss by evaluating PTEN status in only one core. Future studies will assess whether evaluation of PTEN status on all cancer-containing needle cores may increase the sensitivity of our assay for predicting tumor upgrading in Gleason score 6 needle biopsies.
Taken together, our data strongly suggest that there may exist varying ‘molecular’ grades of Gleason pattern 3 prostate cancer, with PTEN protein loss indicating a threefold increased risk of an occult pattern 4 component in the same patient. In combination with increasingly accurate imaging techniques, assessment for molecular markers may help to identify patients at higher risk of harboring more aggressive prostate tumors. PTEN loss, as assessed by immunohistochemistry and perhaps in conjunction with confirmatory FISH, is one such relatively inexpensive marker. Together with a panel of additional markers, it may prove useful to identify patients who are suboptimal candidates for active surveillance protocols.
References
Pound CR, Partin AW, Eisenberger MA et al. Natural history of progression after PSA elevation following radical prostatectomy. JAMA 1999;281:1591–1597.
Ross HM, Kryvenko ON, Cowan JE et al. Do adenocarcinomas of the prostate with Gleason score (GS) </=6 have the potential to metastasize to lymph nodes? Am J Surg Pathol 2012;36:1346–1352.
Pierorazio PM, Walsh PC, Partin AW et al. Prognostic Gleason grade grouping: data based on the modified Gleason scoring system. BJU Int 2013;111:753–760.
Ellis CL, Walsh PC, Partin AW et al. Multiple cores of Gleason score 6 correlate with favourable findings at radical prostatectomy. BJU Int 2013;111:E306–E309.
Berman DM, Epstein JI . When is prostate cancer really cancer? Urol Clin N Am 2014;41:339–346.
Epstein JI, Feng Z, Trock BJ et al. Upgrading and downgrading of prostate cancer from biopsy to radical prostatectomy: incidence and predictive factors using the modified Gleason grading system and factoring in tertiary grades. Eur Urol 2012;61:1019–1024.
Irani J, Blanchet P, Salomon L et al. Is an extended 20-core prostate biopsy protocol more efficient than the standard 12-core? A randomized multicenter trial. J Urol 2013;190:77–83.
Siddiqui MM, Rais-Bahrami S, Truong H et al. Magnetic resonance imaging/ultrasound-fusion biopsy significantly upgrades prostate cancer versus systematic 12-core transrectal ultrasound biopsy. Eur Urol 2013.
Magheli A, Hinz S, Hege C et al. Prostate specific antigen density to predict prostate cancer upgrading in a contemporary radical prostatectomy series: a single center experience. J Urol 2010;183:126–131.
Oh JJ, Hong SK, Lee JK et al. Prostate-specific antigen vs prostate-specific antigen density as a predictor of upgrading in men diagnosed with Gleason 6 prostate cancer by contemporary multicore prostate biopsy. BJU Int 2012;110:E494–E499.
Truong M, Slezak JA, Lin CP et al. Development and multi-institutional validation of an upgrading risk tool for Gleason 6 prostate cancer. Cancer 2013;119:3992–4002.
Capitanio U, Karakiewicz PI, Valiquette L et al. Biopsy core number represents one of foremost predictors of clinically significant gleason sum upgrading in patients with low-risk prostate cancer. Urology 2009;73:1087–1091.
Gershman B, Dahl DM, Olumi AF et al. Smaller prostate gland size and older age predict Gleason score upgrading. Urol Oncol 2013;31:1033–1037.
Freedland SJ, Kane CJ, Amling CL et al. Upgrading and downgrading of prostate needle biopsy specimens: risk factors and clinical implications. Urology 2007;69:495–499.
Moussa AS, Kattan MW, Berglund R et al. A nomogram for predicting upgrading in patients with low- and intermediate-grade prostate cancer in the era of extended prostate sampling. BJU Int 2010;105:352–358.
Lotan TL, Gurel B, Sutcliffe S et al. PTEN protein loss by immunostaining: analytic validation and prognostic indicator for a high risk surgical cohort of prostate cancer patients. Clin Cancer Res 2011;17:6563–6573.
Chaux A, Peskoe SB, Gonzalez-Roibon N et al. Loss of PTEN expression is associated with increased risk of recurrence after prostatectomy for clinically localized prostate cancer. Mod Pathol 2012;25:1543–1549.
Antonarakis ES, Keizman D, Zhang Z et al. An immunohistochemical signature comprising PTEN, MYC, and Ki67 predicts progression in prostate cancer patients receiving adjuvant docetaxel after prostatectomy. Cancer 2012;118:6063–6071.
McMenamin ME, Soung P, Perera S et al. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res 1999;59:4291–4296.
Sircar K, Yoshimoto M, Monzon FA et al. PTEN genomic deletion is associated with p-Akt and AR signalling in poorer outcome, hormone refractory prostate cancer. J Pathol 2009;218:505–513.
Yoshimoto M, Cunha IW, Coudry RA et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer 2007;97:678–685.
Krohn A, Diedler T, Burkhardt L et al. Genomic deletion of PTEN is associated with tumor progression and early PSA recurrence in ERG fusion-positive and fusion-negative prostate cancer. Am J Pathol 2012;181:401–412.
Yoshimoto M, Ludkovski O, DeGrace D et al. PTEN genomic deletions that characterize aggressive prostate cancer originate close to segmental duplications. Genes Chromosomes Cancer 2012;51:149–160.
Yoshimoto M, Ding K, Sweet JM et al. PTEN losses exhibit heterogeneity in multifocal prostatic adenocarcinoma and are associated with higher Gleason grade. Mod Pathol 2013;26:435–447.
Cuzick J, Yang ZH, Fisher G et al. Prognostic value of PTEN loss in men with conservatively managed localised prostate cancer. Br J Cancer 2013;108:2582–2589.
Epstein JI, Allsbrook WC Jr, Amin MB et al. Update on the Gleason grading system for prostate cancer: results of an international consensus conference of urologic pathologists. Adv Anat Pathol 2006;13:57–59.
Sundi D, Kryvenko ON, Carter HB et al. Pathological examination of radical prostatectomy specimens in men with very low risk disease at biopsy reveals distinct zonal distribution of cancer in black American men. J Urol 2014;191:60–67.
Lotan TL, Zhang M, Fedor H et al. ERG and PTEN status of HOXB13 G84E-related prostate cancers: an analysis of 106 cases. Mod Path 2014;27 (Supp 2):246A.
Hong SK, Eastham JA, Fine SW . Localization of higher grade tumor foci in potential candidates for active surveillance who opt for radical prostatectomy. Prostate Int 2013;1:152–157.
Sowalsky AG, Ye H, Bubley GJ et al. Clonal progression of prostate cancers from Gleason grade 3 to grade 4. Cancer Res 2013;73:1050–1055.
Berger MF, Lawrence MS, Demichelis F et al. The genomic complexity of primary human prostate cancer. Nature 2011;470:214–220.
Taylor BS, Schultz N, Hieronymus H et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012;487:239–243.
Barbieri CE, Baca SC, Lawrence MS et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 2012;446:85–89.
Yoshimoto M, Cutz JC, Nuin PA et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet 2006;169:128–137.
Gumuskaya B, Gurel B, Fedor H et al. Assessing the order of critical alterations in prostate cancer development and progression by IHC: further evidence that PTEN loss occurs subsequent to ERG gene fusion. Prostate Cancer Prostatic Dis 2013;16:209–215.
Bostwick DG, Singh H, Hossain D . PTEN losses by fluorescence in situ hybridization (FISH) – but not immunohistochemistry – correlate with higher Gleason Grade: prospective study of 2261 prostate needle biopsies in a community practice setting. Mod Pathol 2013;26:199A.
Reese AC, Landis P, Han M et al. Expanded criteria to identify men eligible for active surveillance of low risk prostate cancer at Johns Hopkins: a preliminary analysis. J Urol 2013;190:2033–2038.
Acknowledgements
Funding for this research was provided in part by a Prostate Cancer Foundation Young Investigator Award (TLL), a grant from David Koch administered by the Patrick C Walsh Prostate Cancer Research Fund (TLL), the NIH/NCI Prostate SPORE P50CA58236, a PhD grant from Fundação para Ciência e Tecnologia (SFRH/ BD/ 69819/ 2010 to FLFC), funds from the Ontario Institute for Cancer Research and Southeast Ontario Academic Medical Organization (to DMB), and Prostate Cancer Canada grant 2012-937 (JAS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
DMB has consulted and/or performed sponsored research for Myriad Genetics and Metamark Genetics; JAS has consulted for CymoGen Dx.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Supplementary information
Rights and permissions
About this article
Cite this article
Lotan, T., Carvalho, F., Peskoe, S. et al. PTEN loss is associated with upgrading of prostate cancer from biopsy to radical prostatectomy. Mod Pathol 28, 128–137 (2015). https://doi.org/10.1038/modpathol.2014.85
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.85
This article is cited by
-
Genomic and phenotypic heterogeneity in prostate cancer
Nature Reviews Urology (2021)
-
High throughput assessment of biomarkers in tissue microarrays using artificial intelligence: PTEN loss as a proof-of-principle in multi-center prostate cancer cohorts
Modern Pathology (2021)
-
Molekularpathologie bei urologischen Tumoren
Der Pathologe (2021)
-
Activation of PAR2 by tissue factor induces the release of the PTEN from MAGI proteins and regulates PTEN and Akt activities
Scientific Reports (2020)
-
Molecular characterization of prostate cancer in Middle Eastern population highlights differences with Western populations with prognostic implication
Journal of Cancer Research and Clinical Oncology (2020)