Abstract
The activation of nuclear factor kappa B (NFκB) transcription factor family is considered to have a key role in diffuse large B-cell lymphoma (DLBCL) pathogenesis and is associated with a specific molecular subtype, the activated B-cell-like (ABC) subtype. We evaluated the expression of NFκB by immunohistochemistry in a large series of DLBCL cases. The five different NFκB family members (NFκB1, NFκB2, RELA, RELB, and REL) showed a heterogeneous expression pattern with the vast majority of cases being positive for at least one factor. Two independent series of tumor samples were classified into germinal center B-cell-like (GCB) or ABC subtypes using different approaches, immunohistochemistry, or gene expression profiling, and the expression of NFκB family members was assessed. Notably, no significant differences regarding the expression of the different NFκB members were detected between the two subtypes, suggesting that NFκB signaling is a prominent feature not only in the ABC subtype, but also in the GCB tumors. Of the five transcription factors, only REL expression had a significant clinical impact on R-CHOP-treated diffuse large B-cell lymphoma, identifying a subgroup of patients with superior clinical outcome.
Similar content being viewed by others
Main
Diffuse large B-cell lymphoma (DLBCL) is the most common type of lymphoma in adults and is clinically and molecularly a heterogeneous disease.1, 2 Based on gene expression profiling, two different molecular subtypes of DLBCL were recognized according to the putative cell of origin; the more aggressive activated B-cell-like (ABC) subtype and the germinal center B-cell-like (GCB) subtype.3, 4, 5 These subtypes reflect distinct differentiation stages of normal B-cell development and differ in their clinical presentation, cure rates, and molecular features.2 Due to practical reasons, algorithms to classify these subtypes using immunohistochemistry of a few markers instead of gene expression profiling have been developed and extensively used.6, 7 These subtypes differ in their oncogenic programs and several subtype-specific genetic alterations have been described.2, 8, 9 A hallmark of the ABC subtype is constitutive nuclear factor kappa B (NFκB) activation, and a disruption of this activation induces apoptosis in ABC cell lines.10 Several recurrent genetic aberrations affecting NFκB signaling have been described in the ABC tumors, including for instance mutations in caspase recruitment domain family member 11, tumor necrosis factor alpha-induced protein 3, CD79, or myeloid differentiation primary response 88.11, 12, 13, 14
The NFκB family consists of five transcription factors: NFκB1 (p50 and its precursor p105), NFκB2 (p52 and its precursor p100), RELA (also called p65), RELB, and REL (also called c-Rel). An activation of the pathway leads to nuclear translocation of NFκB dimers, which are normally held inactive in the cytoplasm in unstimulated cells. Two different pathways using different signaling components are described; the classical pathway that results in nuclear accumulation of dimers containing NFκB1, RELA, and REL, and the alternative pathway, characterized by nuclear accumulation of NFκB2/RELB dimers (reviewed by Vallabhapurapu and Karin15). Even though constitutive NFκB activation is one of the proposed oncogenic mechanisms underlying the molecular pathology of the ABC subtype, the actual expression of NFκB in DLBCL tumor specimens and the presence of both classical and alternative signaling components as well as their clinical correlation have not been well studied.
In this report, we evaluate the expression of the five NFκB transcription factors in human DLBCL tumor biopsies, and analyze their expression pattern, subtype specificity, and clinical impact.
Materials and methods
Patient Samples
The use of human tumor samples in this study was approved by the Clinical Research Ethics Committee of Hospital Universitario Marqués de Valdecilla, HUMV (Santander, Spain). Samples were collected by the tumor banks of the Spanish National Cancer Research Centre (CNIO, Madrid, Spain) and HUMV. Samples were obtained before treatment and were diagnosed according to the current criteria of the World Health Organization (WHO) classification.16 Patient characteristics of a series of 88 DLBCL samples are summarized in Table 2.
Immunohistochemistry, Scoring, and In Situ Hybridization
Formalin-fixed paraffin-embedded tumor tissues were included in duplicates into tissue microarrays and stained as previously described.17 Primary antibodies and immunohistochemistry conditions are listed in Supplementary Table 1. Evaluation and scoring were done by two pathologists (S.M-M and R.S-P). If more than 30% of the tumoral cells presented nuclear staining of a NFκB member, the tumor was considered positive for that marker. This threshold was chosen because it has previously been used for NFκB expression in DLBCL14, 18 and is also a threshold commonly used for standard immunohistochemistry markers in these tumors.6, 7 Tumors positive for Epstein–Barr virus (EBV) determined by the expression of EBV-encoded RNA, EBER, by in situ hybridization (described by Montes-Moreno et al17) were excluded from the study.
DLBCL Subclassification
All tumors were stratified into ABC or GCB subtypes by immunohistochemistry according to the algorithm of Choi,6 based on the expression of CD10, interferon regulatory factor 4, B-cell lymphoma 6, serine peptidase inhibitor, clade A, member 9 (SERPINA9, also called GCET1), and forkhead box protein P1. Sixty samples included in this study had also previously been classified into ABC or germinal B-cell-like DLBCL using gene expression profiling, described by Hu et al.19
Statistical Analyses
All statistical analyses were performed using SPSS software version 15.0 (SPSS, Chicago, IL, USA). Independence of variables was determined using crosstabs applying a Pearson’s χ2-test. The Kaplan–Meier method applying the log-rank test was used to estimate the differences in overall survival between NFκB-positive and negative cases. A multivariate survival analysis was performed using a Cox proportional hazard regression model. Values of P<0.05 were considered statistically significant in all analyses.
Results
Expression Pattern of Different NFκB Members in Diffuse Large B-cell Lymphoma
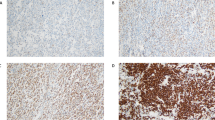
As the family of NFκB consists of five different transcription factors, we decided to evaluate the expression of all of them in order to get a complete picture of the expression pattern in DLBCL. In a series of 113 consecutive human DLBCL biopsies taken at the time of diagnosis, the nuclear expression of NFκB1, NFκB2, RELB, RELA, and REL was assessed (Figure 1a). A tumor was considered positive if more than 30% of its tumoral cells presented nuclear expression (see Materials and Methods section). As a validation of our immunohistochemistry approach, stainings were compared with the DNA binding activity of the different subunits in six DLBCL cell lines, showing that nuclear staining of NFκB corresponded with increased DNA binding of the specific subunits (Supplementary Figure 1). Furthermore, Compagno et al14 showed that nuclear expression of NFκB1 or NFκB2 in DLBCL, evaluated by immunohistochemistry, was associated with enhanced transcription of NFκB target genes, suggesting that immunohistochemical analysis of nuclear NFκB could be a valid approach for assessing active NFκB signaling. The percentages of positive cases for each of the NFκB proteins are represented in Figure 1b. Interestingly, only 12.4% of cases lacked nuclear expression all NFκB subunits. Of the five proteins, REL was the most commonly expressed (64%), followed by RELB (43%), NFκB1 (43%), and NFκB2 (29.2%). Surprisingly, nuclear RELA was only detected in 12% of the samples. Supporting the low frequency of RELA-positive tumors, other studies20, 21, 22 using different antibodies have also reported reduced frequency of RELA expression in DLBCL. The relationship between the expressions of the different subunits is represented in the Venn diagram in Figure 1c. Notably, the expression pattern of the different factors is complex, and does not follow the classical outline of the NFκB pathway. There is a significant correlation between the expression of factors of the classical and alternative pathway, such as between NFκB1 and NFκB2 (χ2-test, P=0.000052), NFκB2 and RELA (P=0.006), and between NFκB1 and RELB (P=0.006), suggesting an overlap between classical and alternative factors (Table 1). It is also remarkable that 23 REL-positive cases do not express any other NFκB factor.
Nuclear factor kappa B (NFκB) expression pattern in diffuse large B-cell lymphoma. (a) Immunohistochemical staining shows nuclear expression of NFκB1, NFκB2, RELB, REL, and RELA in human primary diffuse large B-cell lymphoma samples. (b) Percentages of cases staining positive for NFκB subunits (nuclear staining in >30% of the tumoral cells). NFκB in the last column represents the percentage of cases staining positive for any NFκB subunit. (c) Venn diagram showing the overlap in number of positive cases between the different NFκB factors.
Correlation Between NFκB Expression and DLBCL Subclassification
As the ABC phenotype has been characterized by prominent NFκB activation, we wanted to explore how the expression of the five transcription factors were distributed between the two subtypes. To subclassify the tumors into the molecular subtypes ABC or GCB, we used immunohistochemistry according to the algorithm of Choi6 in a series of 113 DLBCL samples. In this series, 59% of the tumors were classified as ABC and 41% were classified as GCB. Evaluating the nuclear expression of NFκB2, NFκB1, RELB, REL, and RELA in these tumors, we surprisingly saw a fairly even distribution of positive staining between the subtypes. Only a very slight increase of positive cases could be observed in the ABC subgroup, but none of the five transcription factors was significantly differentially expressed between the subtypes (Figure 2a and Table 1). To validate this finding, we performed the study in a second series of 60 DLBCL samples, where subclassification had been done using gene expression profiling (described by Hu et al19). Still, the expression of NFκB (NFκB1 or NFκB2) showed no correlation with molecular subtype (Figure 2b). Moreover, only 5 out of 60 cases were misclassified using immunohistochemistry for subclassification in this series (concordance of 91.7%), suggesting that this is a valid approach for molecular subtype stratification in our cohorts.
Nuclear factor kappa B (NFκB) expression in different molecular diffuse large B-cell lymphoma subtypes. (a) Percentage of positive cases in each diffuse large B-cell lymphoma subtype (white columns represent activated B-cell-like (ABC), and black columns represent germinal center B-cell-like (GCB) tumors. Subclassification was performed using immunohistochemistry according to the algorithm of Choi. (b) Comparison of percentage of positive cases for NFκB1 and NFκB2 in different diffuse large B-cell lymphoma subtypes when classification was performed using gene expression profiling.
Clinical Impact of NFκB in R-CHOP-Treated DLBCL
Samples from a cohort of 88 DLBCL patients treated with R-CHOP immunochemotherapy (rituximab combined with cyclophosphamide, doxorubicin, vincristine, and prednisone) were used to estimate if NFκB expression had any impact on the clinical course of the malignancy (see Table 2 for patient characteristics). Of all the five NFκB subunits, only REL had a significant impact on the clinical outcome. Patients having REL-positive tumors had a significantly superior overall survival compared with patients with REL-negative tumors (Figure 3, P=0.0041). The estimated 5-year overall survival rate was 73.7% in patients with REL-positive tumors compared with 59.7% in patients with REL-negative tumors. In order to take into account other prognostic factors in DLBCL, such as the international prognostic index (IPI) and the molecular subtype, we performed a multivariate analysis including these variables (Supplementary Table 2). This analysis showed that REL was a significant favorable prognostic factor, independent of both the IPI and GCB/ABC classification. The expression of NFκB2 was also associated with enhanced overall survival, although this difference was not statistically significant (Figure 3).
Clinical correlation of nuclear factor kappa B (NFκB) in diffuse large B-cell lymphoma. Kaplan–Meier graphs representing survival curves for patients with NFκB-positive and NFκB-negative tumors. Each plot represents the survival curves of an individual NFκB transcription factor. Numbers of positive and negative cases are written in parenthesis beside each curve. P-values were estimated by a log-rank test. Tumors positive for nuclear REL expression are significantly associated with a superior overall survival compared with REL-negative tumors (P=0.042).
Discussion
There are a few reports assessing the protein expression of NFκB in DLBCL tumors, and to our knowledge, this is the first work reporting the expression of the entire family. Using immunohistochemistry to assess the nuclear expression of the five different transcription factors, we observed an intricate expression pattern. Even though the expression of the different NFκB proteins was heterogeneous in DLBCL, the vast majority of tumors (88%) expressed nuclear NFκB. Correlated expression between different NFκB subunits, such as between NFκB1 and NFκB2, NFκB2 and RELB, NFκB2 and RELA, NFκB1 and RELB, and between NFκB1 and RELA, was observed, although simultaneous expression of other combinations also was present. This remark suggests that the expression pattern of NFκB in tumors does not follow the prototypic NFκB model, and that there is no precise line distinguishing the classical and alternative pathways. However, apart from the most representative and frequent dimerization complexes (NFκB2/RELB, NFκB1/RELA, and NFκB1/REL), both homodimers and dimers including a variety of NFκB members as well as atypical partners have already been described.23 Likewise, a correlation between the expression of classical and alternative NFκB markers has previously been described in T-cell lymphomas.24 Interestingly, REL presented a distinct expression pattern and even though an association between NFκB1 and REL could be perceived (not statistically significant), around a third of the REL-positive tumors did not express any other NFκB member. Yet, nuclear dimers of CD40 and REL have been described to activate transcription of NFκB target genes in DLBCL.25 To determine the different dimerization partners was though beyond the scope of this report. The complicated expression pattern of the different subunits is also an indicator of that looking specifically at only a single NFκB factor as an estimate of NFκB activation will probably lead to a significant underestimation of the actual NFκB signaling.
A hallmark of the ABC subtype is constitutive NFκB activation, and a disruption of this activation induces apoptosis in ABC DLBCL cell lines.10, 26 Our analysis of NFκB expression in a series of 113 DLBCL samples revealed that the majority of the tumors express nuclear NFκB, independent of subclassification. Surprisingly, we found no significant correlation between molecular subtype and the expression of any of the five NFκB members, indicating that NFκB signaling is present not only in the ABC subtype, but also in the GCB subtype. To discard the possibility that this observation could be a result of using immunohistochemistry as a method for classification, we performed the analysis in two independent series of DLBCL tumors, classified both by immunohistochemistry and gene expression profiling. These two analyses showed that both classical and alternative NFκB members were expressed equally in ABC and GCB tumors. In agreement with our observation, previous works analyzing the expression of phosphorylated RELA and NFκB2 as markers for NFκB activation in DLBCL tumors, reported expression of these subunits in both subtypes with similar frequencies.18, 21 On the contrary, other studies associated the expression of nuclear RELA, NFκB1, and NFκB2 with the ABC subgroup, although they were also present in a considerable fraction of the GCB tumors as well.14, 20 An important difference between our study and several others was that EBV-positive cases were excluded from the analysis. Due to their distinct molecular and clinical profile, EBV-positive DLBCL has been suggested as a distinct category of DLBCL.16 These tumors have been characterized by NFκB activation and they frequently present an ABC phenotype.17 Including this category together with EBV-negative diffuse large B-cell lymphoma probably gives rise to bias with an increased number of NFκB-positive cases and a stronger association with the ABC phenotype. Our observation that NFκB signaling is present in both molecular subtypes of DLBCL, evokes several questions concerning whether NFκB is involved in the underlying pathogenesis of the germinal center B-cell-like tumors as well, and whether NFκB inhibition could be useful in a subset of these tumors. As the ABC and GCB subtypes partly present a distinct pattern of genetic abnormalities and respond differently to therapy,2 the mechanisms leading to NFκB signaling and the subsequent transcriptional response might be different in the two subtypes.
There are only a few previous publications describing NFκB signaling and its clinical correlation, and the results are contradictory. In this work, we investigated the impact of all NFκB members on patient overall survival, using nuclear expression as an estimate of NFκB activation. In DLBCL, nuclear REL expression was seen to be a common feature and identified a subset of patients with enhanced overall survival. REL is the only NFκB transcription factor that is frequently amplified in human cancers and has also consistently been shown to have transforming potential in culture.27 Amplification of the REL locus is commonly observed in DLBCL and mainly associated with the GCB phenotype.5 In our study, nuclear REL expression is though detected in both the ABC and the GCB subtypes. However, no correlation between REL amplification and nuclear REL expression was observed in DLBCL in a study by Houldsworth et al.28 NFκB has previously been associated with enhanced patient survival in another DLBCL study, where phosphorylated RELA was assessed.21 On the other hand, RELA expression was described as an adverse prognostic marker in DLBCL in another report.20 Moreover, nuclear REL assessed by immunohistochemistry, was associated with worse clinical outcome in GCB DLBCL in a previous publication.29 Those studies though were performed in biopsies from patients treated with chemotherapy without rituximab. As the addition of rituximab has markedly improved the overall survival rates in DLBCL, and the outcome of survival analyses depends on the clinical management, this can be a possible explanation for the observed differences. Our study is performed using a cohort of patients that has uniformly been treated with R-CHOP, which is the standard treatment for DLBCL patients nowadays. The fact that only REL expression has prognostic impact on DLBCL suggests that different NFκB subunits have different roles and activate different transcriptional programs. However, the roles of specific subunits or NFκB dimers regulating transcriptional selectivity, are far from understood and await further research.
References
Coiffier B, Thieblemont C, Van Den Neste E et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d'Etudes des Lymphomes de l'Adulte. Blood 2010;116:2040–2045.
Lenz G, Staudt LM . Aggressive lymphomas. N Engl J Med 2010;362:1417–1429.
Alizadeh AA, Eisen MB, Davis RE et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–511.
Wright G, Tan B, Rosenwald A et al. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A 2003;100:9991–9996.
Rosenwald A, Wright G, Chan WC et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 2002;346:1937–1947.
Choi WW, Weisenburger DD, Greiner TC et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res 2009;15:5494–5502.
Hans CP, Weisenburger DD, Greiner TC et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004;103:275–282.
Pasqualucci L, Trifonov V, Fabbri G et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 2011;43:830–837.
Zhang J, Grubor V, Love CL et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci 2013;110:1398–1403.
Davis RE, Brown KD, Siebenlist U et al. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med 2001;194:1861–1874.
Davis RE, Ngo VN, Lenz G et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010;463:88–92.
Lenz G, Davis RE, Ngo VN et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 2008;319:1676–9.
Ngo VN, Young RM, Schmitz R et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011;470:115–119.
Compagno M, Lim WK, Grunn A et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 2009;459:717–721.
Vallabhapurapu S, Karin M . Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 2009;27:693–733.
Swerdlow SH, Campo E, Harris NL et al. WHO classification of tumours of haematopoietic and lymphoid tissue 4th edn. WHO Press IARC: Lyon, 2008, pp 233–237.
Montes-Moreno S, Odqvist L, Diaz-Perez JA et al. EBV-positive diffuse large B-cell lymphoma of the elderly is an aggressive post-germinal center B-cell neoplasm characterized by prominent nuclear factor-kB activation. Mod Pathol 2012;25:968–982.
Pham LV, Fu L, Tamayo AT et al. Constitutive BR3 receptor signaling in diffuse, large B-cell lymphomas stabilizes nuclear factor-kappaB-inducing kinase while activating both canonical and alternative nuclear factor-kappaB pathways. Blood 2011;117:200–210.
Hu S, Xu-Monette ZY, Balasubramanyam A et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood 2013;121:2715–2724.
Bavi P, Uddin S, Bu R et al. The biological and clinical impact of inhibition of NF-kappaB-initiated apoptosis in diffuse large B cell lymphoma (DLBCL). J Pathol 2011;224:355–366.
Espinosa I, Briones J, Bordes R et al. Activation of the NF-kappaB signalling pathway in diffuse large B-cell lymphoma: clinical implications. Histopathology 2008;53:441–449.
Talwalkar SS, Valbuena JR, Abruzzo LV et al. MALT1 gene rearrangements and NF-kappaB activation involving p65 and p50 are absent or rare in primary MALT lymphomas of the breast. Mod Pathol 2006;19:1402–1408.
Hoffman A, Natoli G, Ghosh G . Circuitry of nuclear factor kappaB signaling. Immunol Rev 2006;210:171–186.
Odqvist L, Sanchez-Beato M, Montes-Moreno S et al. NIK controls classical and alternative NF-kappaB activation and is necessary for the survival of human T-cell lymphoma cells. Clin Cancer Res 2013;19:2319–2330.
Zhou HJ, Pham LV, Tamayo AT et al. Nuclear CD40 interacts with c-Rel and enhances proliferation in aggressive B-cell lymphoma. Blood 2007;110:2121–2127.
Lam LT, Davis RE, Pierce J et al. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res 2005;11:28–40.
Gilmore TD, Kalaitzidis D, Liang MC et al. The c-Rel transcription factor and B-cell proliferation: a deal with the devil. Oncogene 2004;23:2275–2286.
Houldsworth J, Olshen AB, Cattoretti G et al. Relationship between REL amplification, REL function, and clinical and biologic features in diffuse large B-cell lymphomas. Blood 2004;103:1862–1868.
Curry CV, Ewton AA, Olsen RJ et al. Prognostic impact of C-REL expression in diffuse large B-cell lymphoma. J Hematop 2009;2:20–26.
Acknowledgements
Red Temática de Investigación Cooperativa en Cáncer (RTICC; RD06/0020/0107, RD012/0036/0060), Asociación Española Contra el Cáncer (AECC), MINECO (SAF2008-03871), and FIS (FIS-PI10/00621) provided financial support for this work. M.S-B is supported by a Miguel Servet-contract (FIS-CP11/00018). The authors thank María-Jesus Artiga for sample administration and Francisco X. Real for scientific discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Modern Pathology website
Rights and permissions
About this article
Cite this article
Odqvist, L., Montes-Moreno, S., Sánchez-Pacheco, R. et al. NFκB expression is a feature of both activated B-cell-like and germinal center B-cell-like subtypes of diffuse large B-cell lymphoma. Mod Pathol 27, 1331–1337 (2014). https://doi.org/10.1038/modpathol.2014.34
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2014.34
Keywords
This article is cited by
-
Cell-type-specific sensitivity of bortezomib in the methotrexate-resistant primary central nervous system lymphoma cells
International Journal of Clinical Oncology (2019)
-
HDAC6 regulates microRNA-27b that suppresses proliferation, promotes apoptosis and target MET in diffuse large B-cell lymphoma
Leukemia (2018)
-
The role of bortezomib in newly diagnosed diffuse large B cell lymphoma: a meta-analysis
Annals of Hematology (2018)
-
Low dose of lenalidmide and PI3K/mTOR inhibitor trigger synergistic cytoxicity in activated B cell-like subtype of diffuse large B cell lymphoma
Journal of Experimental & Clinical Cancer Research (2016)
-
Diagnostic and predictive biomarkers for lymphoma diagnosis and treatment in the era of precision medicine
Modern Pathology (2016)