Abstract
Overexpression of the BCL2 protein has been reported as a poor prognostic factor for diffuse large B-cell lymphoma (DLBCL). However, there are currently no standardized criteria for evaluating BCL2 protein expression. We aimed to evaluate the prognostic value of BCL2 expression determined by immunohistochemistry (IHC), incorporating both the staining intensity and proportion, in patients with de novo DLBCL who received rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) as first-line treatment. We defined tumors with BCL2 expression in nearly all tumor cells with a uniformly strong intensity by IHC as BCL2 super-expressor. The BCL2 super-expressors (n = 35) showed significantly worse event-free survival (EFS; HR, 1.903; 95% CI, 1.159–3.126, P = 0.011) and overall survival (OS; HR, 2.467; 95% CI, 1.474–4.127, P = 0.001) compared with the non-BCL2 super-expressors (n = 234) independent of the international prognostic index (IPI), cell of origin (COO), and double expressor status in the training set (n = 269). The adverse prognostic impact of BCL2 super-expression was confirmed in the validation set (n = 195). When the survival outcomes were evaluated in the entire cohort (n = 464), BCL2 super-expressor group was significantly associated with inferior EFS and OS regardless of IPI, COO, MYC expression, and stages. BCL2 super-expressors had genetic aberrations enriched in the NOTCH and TP53 signaling pathways. This study suggests that the BCL2 super-expressor characterizes a distinct subset of DLBCL with a poor prognosis and warrants further investigation as a target population for BCL-2 inhibitors.
Similar content being viewed by others
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of non-Hodgkin lymphoma. It is a heterogeneous disease characterized by distinct pathologic subtypes, gene expression profiles, and genetic alterations. The addition of rituximab (R), an anti-CD20 monoclonal antibody, to CHOP (cyclophosphamide, doxorubicin, vincristine, prednisone) has significantly improved the survival outcomes in patients with DLBCL. However, even with R-CHOP therapy, up to 40% of the patients are refractory to treatment or eventually experience a relapse. The survival outcomes in those patients are very poor despite salvage therapy1,2,3. Therefore, it is crucial to identify DLBCL subgroups with a poor prognosis, which may provide opportunities to optimize the treatment strategies for these patients.
During the last two decades, numerous efforts have been made to identify unique subtypes of DLBCL. Gene expression profiling studies have classified DLBCL into two distinct subgroups based on the cell of origin (COO) and revealed that activated B-cell-type (ABC) DLBCL has a poor survival outcome compared with germinal center B-cell-type (GCB) DLBCL4,5. Several trials have been conducted to improve the survival of patients with ABC or non-GCB DLBCL by combining novel agents to R-CHOP6,7,8. However, all of these trials have failed to demonstrate a significant survival benefit. Different genetic subtypes of DLBCL have recently been identified based on comprehensive genetic analysis using next-generation sequencing (NGS) techniques9,10. Nevertheless, NGS modalities have not yet been adopted into routine clinical practice, and tailored treatment based on different genomic subtypes has not, however, been clearly defined.
Another approach for subtyping DLBCL is based on its chromosomal rearrangement. Concurrent rearrangements involving MYC along with BCL2 and/or BCL6 are defined as double-hit or triple-hit lymphomas (DH/THLs) and comprise approximately 2–10% of newly diagnosed patients with DLBCL11. Also, coexpression of MYC and BCL2 proteins with/without underlying rearrangements is considered another adverse prognostic indicator termed double-expressor lymphoma (DEL)12,13,14. Nevertheless, robust prospective data validating the role of more effective treatments in DH/THLs and DEL are lacking.
Recently, there has been growing interest in BCL2 with the introduction of venetoclax, a highly selective and potent BCL2 inhibitor, which demonstrated promising efficacy, particularly in BCL2 immunohistochemistry (IHC) positive patients with DLBCL15. Thus, an accurate and prompt decision on the BCL2 expression status is becoming more important for treatment decisions as well as prognostication. The BCL2 is an oncogene located on chromosome 18q21 and encodes a pro-survival protein that maintains cellular viability by inhibiting the mitochondrial apoptotic pathway. The overexpression of the BCL2 protein has been reported to be associated with resistance to chemotherapy16,17. In previous studies, the proportion of patients with BCL2 IHC positive DLBCL was highly variable, ranging from approximately 40–80%13,14,18. This discrepancy was mainly due to the various definitions of positivity with varying cut-off values, as there are no standardized criteria. Consequently, the prognostic impact of BCL2 protein expression varies between studies. More recently, a study reported that uniformly strong BCL2 expression intensity in >90% of the tumor cells was independently associated with poor survival outcome, irrespective of MYC expression, in DLBCL patients treated with R-CHOP19. However, its underlying genetic features have not been evaluated. We postulated these patients with a high percentage of cells showing a vigorous BCL2 staining intensity by IHC might constitute a distinct subgroup of DLBCL with distinct genetic features.
This study aimed to evaluate the prognostic value of BCL2 expression determined by IHC, incorporating both the staining intensity and proportion, in patients with de novo DLBCL who received R-CHOP as first-line treatment. In addition, we evaluated the underlying oncogenic pathways resulting in BCL2 deregulation in patients with BCL2 expression in nearly all tumor cells with a uniformly strong intensity by IHC.
Methods
Patients
We retrospectively collected formalin-fixed paraffin-embedded (FFPE) diagnostic biopsies from 269 patients with de novo DLBCL between February 2007 and December 2012 at Asan Medical Center (n = 185) and between January 2006 and December 2012 at Ajou University Hospital (n = 84), South Korea. Tissues obtained by core needle biopsy were excluded. All patients underwent standard staging procedures including bone marrow aspiration and biopsy, computed tomography (CT) scans, and positron emission tomography-CT, and received R-CHOP as first-line treatment. Clinical data were obtained by reviewing medical records, and the baseline data collected included age, sex, lactate dehydrogenase levels, Ann Arbor stage, performance status (PS) according to the Eastern Cooperative Oncology Group (ECOG) scale, International Prognostic Index (IPI), and COO based on Hans classification20. Those without appropriate clinical information and/or the histologic materials were excluded. An independent validation set included 195 patients diagnosed with de novo DLBCL between August 2013 and June 2018 at Asan Medical Center, South Korea. All patients in the validation set also received first-line R-CHOP. A subset of patients in the current study was included in a previous study21. A tissue microarray (TMA) comprising at least two representative 1.5 mm cores from each tumor was constructed. All retrieved cases were comprehensively reviewed, and patients with insufficient tumor tissue for constructing the TMA were excluded from the study cohort. This study was approved by the institutional review boards of the participating institutions.
Immunohistochemistry (IHC)
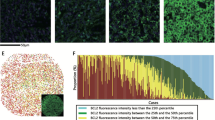
All tissues were fixed in 10% neutral buffered formalin at least for 12 h. IHC for BCL2 (1:100, clone 124, Dako, Glostrup, DK) and MYC (1:50, clone Y69, Abcam, Cambridge, UK) were performed as described previously21. Briefly, 4 μm sections were mounted on silanized charged slides and allowed to dry for 10 min at room temperature and then for 20 min at 65 °C. After deparaffinization, heat-induced epitope retrieval using standard Cell Conditioning 1 was performed for 24 min. The primary antibodies were labeled using the automated immunostaining system with the OptiView DAB Detection Kit (Ventana Medical Systems, USA). BCL2 IHC slides were evaluated by two expert hematopathologists independently (C-S.P. and J.R.). In case of discordance, a discussion for consensus was held using a multi-headed microscope. BCL2 was evaluated in terms of the proportion and intensity of expression. Considering variation between samples and batches, we analyzed the staining intensity of BCL2 expression by comparing it with adjacent reactive T-cells as an internal control. BCL2 expression was classified into the following four categories: (1) score 0: totally negative or a few (<50%) weak positive tumor cells; (2) score 1: the majority of tumor cells (>50%) showing diffuse weak expression; (3) score 2: the majority of tumor cells (>50%) showing heterogeneous or diffuse moderate to strong expression; (4) score 3: nearly all tumor cells showing intense expression. Specifically, tumors with a BCL2 score of 3 uniformly showed a BCL2 expression intensity as strong or stronger than that of adjacent reactive T cells (Fig. 1). We defined tumors with a BCL2 score of 3 as BCL2 super-expressors. In addition, as H-score is a well-known scoring system incorporating staining intensity and proportion of a protein expression, we further evaluated BCL2 super-expressor using H-score in a subpopulation of patients in the training set (n = 184). The details of the H-score evaluation are described in the Supplementary Methods. Moreover, the coefficient of variation of BCL2 H-score among TMA cores of non-BCL2 super-expressors and BCL2 super-expressors was assessed to evaluate the heterogeneity in BCL2 expression among TMA cores.
A BCL2 score 0 shows no BCL2 expression in almost all tumor cells or weak BCL2 expression in very few cells. B BCL2 score 1 shows weak or moderate-intensity BCL2 expression in a subset of tumor cells (<50%). C BCL2 score 2 shows moderate or higher intensity BCL2 expression in most tumor cells (>50%), but the expression pattern is heterogeneous. D BCL2 score 3 shows uniformly strong BCL2 expression in almost all tumor cells.
Cytogenetic analysis
According to the manufacturer’s instructions, fluorescent in situ hybridization (FISH) analyses for BCL2 and MYC were performed on the TMA sample. The Vysis LSI MYC Dual Color Break Apart Rearrangement Probe and the Vysis LSI BCL2 Dual Color Break Apart Rearrangement Probe (Abbot Molecular, Abbot Park, IL, US) were used. The paraffin sections were prepared using a VP 2000 FISH (Abbot Molecular, Abbot Park, IL, US) and the reagents supplied with the kit. After the FISH probes were added to the section, the denaturation and hybridization steps were performed. Nuclei were counterstained with 4’6-diamidino-2-phenylindole (DAPI). For the evaluation, a minimum of 100 cells with clear non-overlapping borders were selected. In a normal diploid cell, two sets of orange/green-fused signals were visible. A sample was considered positive for the rearrangement if >15% of the nuclei exhibited a break-apart signal22. The presence of more than four copies of BCL2 was considered as copy number gain (BCL2GA)23.
BCL2 mRNA quantification
The abundance of BCL2 mRNA was quantified by the NanoString assay. FFPE samples were cut into 10 µm sections for RNA isolation. Total RNA was isolated using the MasterPure Complete DNA/RNA Purification Kit (Lucigen, WI, USA). The nanoString nCounter human mRNA expression assay (nanoString Technologies, WA, USA) was performed with 1 µg of total RNA. Hybridization was carried out by combining each RNA sample with nCounter Reporter probes in hybridization buffer and nCounter Capture probes. The abundance of specific target molecules was quantified on the nCounter Digital Analyzer by counting the individual fluorescent barcodes and assessing the target molecules. Target mRNA expression was normalized to the housekeeping genes (ISY1, TRIM56, UBXN4) and analyzed on nSolver v4.0 (nanoString Technologies).
Targeted NGS and data analysis
We performed targeted NGS for 95 genes selected based on previous literature24,25,26,27. The sequenced genes can be grouped into the following 14 pathways: B-cell development and differentiation, B-cell receptor and Toll-like receptor signaling, NF-κB pathway, MAPK-ERK pathway, PI3K-AKT-mTOR pathway, p53 and DNA damage, cell cycle, apoptosis, NOTCH pathway, cell migration, JAK-STAT pathway, epigenetic regulators, immune escape, and others. The specific genes are listed in Supplementary Table 1.Genomic DNA was extracted using the MasterPure Complete DNA/RNA Purification Kit (Lucigen). The yield and purity of the extracted DNA were assessed using the Nanodrop 2000 Spectrophotometer (Thermo Fisher Scientific, MS, USA). After evaluation of DNA quality, further experiments were performed on 31 of the 35 BCL2 super-expressor samples. For each sample, 50–200 ng of genomic DNA was sheared using a Covaris LE220 focused ultrasonicator (Covaris, Woburn, MA, US). Capture-libraries were assessed for size distribution and quality, and then sequenced on an Illumina HiSeq 2500. The average read depth across all the samples was 1,137x reads per base.
The raw reads in FASTQ file format were initially trimmed for a low-quality base by ngsShoRT28. The trimmed sequences were mapped to the reference human genome (GRCh37) using BWA-MEM29. Using the Picard tool, read group information regarding the genomic coordinates for the sorting and duplicate markings were added to the aligned sequences. The variant calling and filtration for somatic mutations were carried out with GATK MuTect230. The functions of each variant and population statistics were annotated using Annovar31. Unmapped reads, PCR duplicates, and off-target variants were excluded. In total, 3,922 mutations that alter protein function were detected (missense, nonsense, frameshift insertion/deletion, inframe insertion/deletion, nonstop, and splice site). Several filtering steps were applied to obtain meaningful somatic mutations10. The initial inclusion criteria were (1) variant read count ≥ 3 and variant allele frequency ≥ 0.1; (2) missense variants and truncating variants. The exclusion criteria were: (1) variants listed in dbSNP (version 150); (2) variants with a population frequency > 0.0001 in the ExAC database (release 2015); (3) variants that were presumed to be errors or artifacts by the analysis pipeline (GATK tool FilterMutectCall)32. Finally, variants known to be important somatic mutations for cancer were re-included through manual curation. Additionally, we compared the mutation profile of the BCL2 super-expressor with that of DLBCL NOS from publicly available resources9,33.
Statistical analysis
Overall survival (OS) was calculated from diagnosis until death from any cause or was censored at last follow-up. Event-free survival (EFS) was defined as the time from the date of diagnosis till death from any cause, tumor progression or relapse, or the initiation of subsequent anti-lymphoma therapy. Survival rates and corresponding standard errors were estimated using the Kaplan–Meier method, and survival curves were compared using the log-rank test. Baseline characteristics of the groups were compared using Pearson’s chi-square test or Fisher’s exact test for categorical variables and Student’s t-test or Mann–Whitney U-test for continuous variables, as appropriate. The concordance rate for discriminating the BCL2 super-expressors between the two hematopathologists (J.R and C-S.P) was evaluated by the Kappa score.
Univariate and multivariate analyses were performed using Cox proportional hazards regression modeling, and the results are presented as estimated hazard ratios (HRs) with 95% confidence intervals (CIs). Key baseline characteristics and candidate prognostic factors, including sex, IPI, COO, DEL (BCL2 IHC expression ≥50% and MYC IHC expression ≥ 40%), and BCL2 super-expressor status were included in the univariate analysis. In the multivariate analysis, variables exhibiting a significant association with survival (P < 0.2) in the univariate analysis were included, and a two-sided P < 0.05 was considered statistically significant. All statistical analyses were performed using R version 3.6.0. (R Foundation for Statistical Computing, https://www.R-project.org/).
Results
Clinicopathological characteristics of patients in the training set
The baseline characteristics of the patients in the training set (n = 269) are presented in Table 1. The median age was 58 years (range, 48–68), 153 patients (56.9%) were male, and 82 (30.5%) were DELs. Thirty-five patients (13.0%) were classified as BCL2 super-expressors. The baseline characteristics were similar between the BCL2 super-expressor (n = 35) and non-BCL2 super-expressor (n = 234) groups. A higher proportion of patients in the BCL2 super-expressor group had high BCL2 expression (100% vs. 60.7%, P value < 0.001) based on the conventional cut-off (BCL2 positive cells ≥ 50%), had BCL2GA (74.2% vs. 5.3%, P value < 0.001). The reproducibility to classify tumors as BCL2 super-expressor was excellent, with almost perfect interobserver agreement (Kappa = 0.967) between the two hematopathologists (J.R, C-S.P). In contrast, the agreement to classify tumors according to the conventional cut-off (proportion of BCL2 positive cells ≥ 50%) was moderate (Kappa = 0.661).
H-score was evaluable in 184 patients in the training set. BCL2 super-expressor had a significantly higher H-score than non-BCL2 super-expressor (P value < 0.001; Supplementary Fig. 1A). Nineteen patients were classified as extremely high H-score cases (H-score of ≥207.46 [90th percentile]) (Supplementary Fig. 1B), and there was a 92.5% agreement between H-score (≥90th percentile vs. <90th percentile) and BCL2 super-expressor status (Supplementary Table 2). In addition, the coefficient of variation of H-score was significantly lower among TMA cores of BCL2 super-expressor than non-BCL2 super-expressor (P value < 0.001; Supplementary Fig. 1C).
BCL2 super-expressor status and survival outcome in the training set
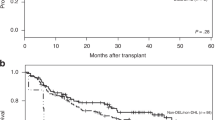
Next, we examined the association of the BCL2 super-expressors with survival outcomes in the training set. With a median follow up duration of 122.0 months (95% CI, 118.0–1), the five-year EFS rate was 64.0% (95% CI, 58.5–70.0) and the five-year OS rate was 70.6% (95% CI, 65.4–76.3) for entire patients in the training set. The BCL2 super-expressor group had significantly worse EFS compared with the non-BCL2 super-expressor group, with five-year EFS rates of 42.2% (95% CI, 28.5–62.5) and 67.2% (95% CI, 61.4–73.5), respectively (P = 0.003) (Fig. 2A). Similarly, patients in the BCL2 super-expressor group demonstrated a significantly worse OS than those in the non-BCL2 super-expressor group, with five-year OS rates of 45.7% (95% CI, 31.9–65.6) and 74.4% (95% CI, 69.0–80.2), respectively (P < 0.001) (Fig. 2B).
Univariate and multivariate analyses of survival outcomes in the training set
Univariate and multivariate analyses of the potential prognostic factors for EFS and OS were performed on the training set. In the univariate analysis, male, high IPI (score of 3–5), non-GCB group, double expressor, MYC expression of ≥40% (IHC), and BCL2 super-expressor group showed potential associations (P < 0.2) with both poor EFS and OS (Table 2 and Supplementary Table 3). Multivariate analyses incorporating double expressor status or MYC expression (IHC) were performed separately due to multicollinearity between these two variables. In the multivariate analysis incorporating double expressor status, the BCL2 super-expressor group remained as an independent prognostic factor for both poor EFS (vs. non-BCL2 super-expressor group; HR, 1.903; 95% CI, 1.159–3.126, P = 0.011) and OS (vs. non-BCL2 super-expressor group; HR, 2.467; 95% CI, 1.474–4.127, P = 0.001) (Table 2). High IPI (score of 3–5) and non-GCB type were also independently associated with EFS and OS. Similar findings were observed in the multivariate analysis incorporating MYC expression (IHC); the BCL2 super-expressor group remained as an independent prognostic factor for poor EFS and OS (Supplementary Table 3).
Validation of BCL2 super-expressor in the validation set
The adverse prognostic impact of BCL2 super-expression was further validated in the independent validation set (n = 195). The patient characteristics of the validation set are described in Supplementary Table 4. Thirty-two patients (16.4%) were classified as BCL2 super-expressors, and the BCL2 super-expressor group had significantly poor EFS and OS compared with the non-BCL2 super-expressor group. The five-year EFS and OS rates between the BCL2 super-expressor and the non-BCL2 super-expressor groups were 43.3 (95% CI, 29.0–64.6) vs. 70.9% (95% CI, 64.2–78.3) (P = 0.004) and 49.3 (95% CI, 34.5–70.4) vs. 72.5% (95% CI, 65.8–79.8) (P = 0.016), respectively (Fig. 3A, B).
(A, B) EFS and OS of patients with low IPI (0–2). (C, D) EFS and OS of patients with high IPI (3–5). (E, F) EFS and OS of patients with GCB type DLBCL. (G, H) EFS and OS of patients with non-GCB type DLBCL. (I, J) EFS and OS of patients with stages 1–2. (K, L) EFS and OS of patients with stages 3–4.
Survival outcomes according to BCL2 super-expressor status within the IPI, COO subtype, stage, and DEL patient subgroups
We further investigated the survival outcomes according to the BCL2 super-expressor status within the IPI, COO subtype, stage, and DEL patient subgroups in the entire cohort (n = 464). BCL2 super-expressor status was significantly associated with inferior EFS and OS within both low IPI (score of 0–2) and high IPI (score of 3–5) groups (P < 0.05; Fig. 3A–D); within both GCB and non-GCB subtypes (P < 0.05; Fig. 3E–H); and within both stage 1–2 and stage 3–4 (P < 0.05; Fig. 3I–L) subgroups. In addition, the BCL2 super-expressor status was also associated with inferior EFS and OS within the non-DEL group (P < 0.001; Supplementary Fig. 2A, B). However, there were no significant differences in EFS and OS according to the BCL2 super-expressor status within the DEL group (P = 0.920; Supplementary Fig. 2C, D).
BCL2 GA and survival outcome
We evaluated the association between BCL2 super-expressor status and BCL2GA and the prognostic value of BCL2GA in the training set. A total of 219 patients were evaluable for BCL2GA; BCL2GA was observed in 33 patients (15.1%). Among the patients with BCL2GA, 23 patients (69.7%) were BCL2 super-expressors, while among the patients without BCL2GA, 8 (4.3%) were BCL2 super-expressors (P < 0.001). There was a significant difference in the survival outcome according to the BCL2GA status. The five-year EFS rate was 48.0% (95% CI, 33.5–68.7) in patients with BCL2GA compared with 68.4% (95% CI, 62.0–75.5) for patients without BCL2GA (P = 0.017) and the corresponding five-year OS rate was 51.5% (95% CI, 37.0–71.7) vs. 81.1% (95% CI, 69.9–82.2), respectively (P = 0.004) (Supplementary Fig. 3A, B). To analyze whether the negative prognostic effect of BCL2 super-expression was dependent on BCL2GA, we evaluated the survival outcomes according to BCL2GA and the BCL2 super-expressor status. While patients with BCL2 super-expression/BCL2GA (+/−) and (+/+) demonstrated significantly worse EFS compared with BCL2 super-expression/BCL2GA (−/−) patients, there were no significant differences in EFS between patients with BCL2 super-expression/BCL2GA (+/−) and (−/−) (Supplementary Fig. 3C). Patients with BCL2 super-expression/BCL2GA (+/−), (+/−), and (+/+) were all associated with significantly worse OS compared with BCL2 super-expression/BCL2GA (–/–) patients (Supplementary Fig. 3D).
Association of BCL2 IHC and BCL2 GA with BCL2 mRNA expression
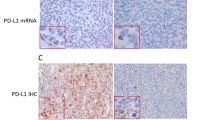
We next evaluated the association between BCL2 mRNA expression levels and BCL2 IHC in 125 patients who were evaluable for BCL2 mRNA expression levels in the training set. Patients with BCL2 super-expression (n = 13) demonstrated significantly higher BCL2 mRNA expression levels compared with patients with high BCL2 expression according to the conventional cut-off value (BCL2 IHC ≥ 50%, n = 69), and those with low BCL2 expression (BCL2 IHC < 50%, n = 43) (P < 0.05; Supplementary Fig. 4A). We further evaluated the association between BCL2 mRNA expression and BCL2GA in 109 patients who were evaluable for both BCL2 mRNA expression and BCL2GA. Patients with BCL2GA (n = 15) was associated with significantly higher BCL2 mRNA expression levels compared to those with no BCL2GA (n = 94) (P < 0.001; Supplementary Fig. 4B)
Genetic features of BCL2 super-expressors
Targeted NGS was performed to evaluate the genomic features of the BCL2 super-expressors. Of the 35 BCL2 super-expressor samples, 31 could be sequenced, and a total of 403 non-synonymous alterations were identified. Of these, the most frequent alterations were missense mutations (n = 251, 62.3%), followed by frameshift deletion (n = 53, 13.2%), nonsense mutation (n = 40, 9.9%), and frameshift insertion (n = 22, 5.5%), respectively. The most frequently mutated genes were KMT2D (14/31 cases, 45.2%), followed by TP53 (13/31 cases, 41.9%), HIST1H2BC (11/31 cases, 35.5%), CD79B (9/31 cases, 29.0%), PIM1 (9/31 cases, 29.0%), EP300 (7/31 cases, 22.6%), PRDM1 (6/31 cases, 19.4%), SPEN (6/31 cases, 19.4%), TMSB4X (6/31 cases, 19.4%), and CREBBP (5/31 cases, 16.1%) (Fig. 4A). Mutation of the BCL2 gene was identified in one case and is detailed in Supplementary Table 5.
A The mutational heatmap visualizes the mutational profile of the frequently mutated top 30 genes and BCL2. The top barplot represents the mutation frequency for each sample, and the right bar plot represents the mutation frequency for each gene. Mutation data are sorted according to the cell of origin. B Enriched oncogenic pathway in BCL2 super-expressor is plotted.
The genetic characteristics of BCL2 super-expressors were compared with those previously reported in unselected DLBCL patients9,33. BCL2 super-expressors showed enrichment of the NOTCH and TP53 signaling pathways (Fig. 4B). In addition, mutations associated with epigenetic regulation such as HIST1H2BC and EP300 were more frequently observed in BCL2 super-expressors (Supplementary Fig. 5). The mutation profiles of the BCL2 super-expressors did not match with any of the previously reported molecular subgroups9,10,34.
Discussion
Considering the failure of clinical trials to improve treatment outcomes based on the cell of origin or double expressor status, it is crucial to identify a more discrete aggressive subgroup representing more homogeneous biology with robust selection criteria. In this regard, the clinical importance of BCL2 is on the rise following the recent introduction of a highly potent BCL2 inhibitor, and its prognostic and predictive value in DLBCL is being actively investigated. This study evaluated the prognostic impact of BCL2 super-expressor status, defined as BCL2 expression in nearly all tumor cells with a uniformly strong intensity as determined by IHC, in newly diagnosed DLBCL patients who were treated with first-line R-CHOP chemoimmunotherapy. The BCL2 super-expression was significantly associated with inferior EFS and OS, and the findings were validated in the independent cohort. Furthermore, BCL2 super-expression stratified the survival outcomes within patients in the low or high IPI, stage 1–2 or 3–4, GCB or non-GCB, and non-DEL groups. On the other hand, there were no significant differences in survival outcomes according to BCL2 super-expressor status in patients with DEL. Given that the biology of DEL is still unclear, despite its adverse prognostic impact, whether the BCL2 super-expressors represent a distinct subgroup within DEL needs further investigation.
The prognostic impact of BCL2 over-expression in patients with DLBCL has been explored in several studies, with contrasting results13,14,18. Although some studies have demonstrated the association between BCL2 over-expression and poor survival18,35,36, others reported that BCL2 over-expression alone might not significantly impact patient survival13,14. One of the reasons for this discrepancy is the different cut-off values used in each study to determine BCL2 IHC positivity. In addition, most previous studies only evaluated the BCL2 expressing cell proportion regardless of the staining intensity. Using only the proportion of stained cells to determine protein expression status can cause discrepancies in the interpretation between pathologists37,38. Indeed, the interobserver agreement for determining high BCL2 expression according to the conventional cut-off value, defined as the proportion of BCL2 positive cells ≥ 50%, was moderate. In comparison, the BCL2 super-expressor status was based on clear criteria that incorporated both the staining intensity and high cut-off value for the proportion of stained cells. Thus, it showed an excellent interobserver agreement (Kappa = 0.967). In addition, we demonstrated that the H-score of the BCL2 super-expressor was significantly higher than that of the non-BCL2 super-expressor. Thus, there was a good agreement between H-score and BCL2 super-expressor status. Considering that assessing H-score is complicated and too cumbersome to employ in clinical practice, assessing BCL2 super-expressor status would be a more practical and effective method in evaluating the expression pattern and intensity of BCL2.
The phase II CAVALLI trial demonstrated the addition of venetoclax to R-CHOP might improve BCL2 positive DLBCL patients’ survival outcomes, suggesting the importance of appropriate biomarkers for targeted therapy15. Considering that the BCL2 protein levels may indicate how dependent a cell is on BCL2 for survival39, we suppose further investigation is necessary to explore the role of BCL2 super-expression as a predictive biomarker for BCL2 inhibitors.
A recent study demonstrated that the BCL2GA is associated with higher levels of BCL2 mRNA expression and increased BCL2 protein expression by IHC40. In line with these results, BCL2GA was significantly correlated with high BCL2 mRNA levels as wells as BCL2 super-expression in the current study. This suggests that BCL2GA is one of the oncogenic pathways that result in BCL2 deregulation, which translates to higher levels of BCL2 mRNA expression and BCL2 protein expression. In addition, BCL2GA was associated with poor survival outcomes, which is consistent with the findings of previous studies41,42. However, there were no significant differences in the EFS according to BCL2GA status in non-BCL2 super-expressors. This result indicates that BCL2 super-expression has a better prognostic value than BCL2GA, although this needs to be further validated in an independent cohort of patients. The better prognostic value of BCL2 super-expression may be related to the fact that the protein expression likely represents an ultimate measure of a particular gene’s activity43.
We further analyzed the molecular characteristics of the BCL2 super-expressor. At the DNA level, a BCL2 exon mutation was only observed in one BCL2 super-expressor patient in the current study, which is much less than previously reported in unselected DLBCL patients9,10. This implies that the function of the intact BCL2 protein may contribute to the biology of the BCL2 super-expressor. Hence BCL2 can be a candidate target for the treatment. Of note, mutations related to the NOTCH and TP53 signaling pathways were frequently observed in the BCL2 super-expressor. The NOTCH signaling pathway is known to be associated with cell proliferation, differentiation, survival, and apoptosis, and TP53 mutation is related to the failure of a cell to undergo cell cycle arrest, DNA repair, and apoptosis44. Recently, based on the advancement of in-depth genomic analyses, new genetic subtypes of DLBCL have been proposed, designated as the LymphGen classification and DLBCL clusters10,45. The BN2 and A53 subtype of LymphGen classification and the C1 and C2 clusters of DLBCL are associated with NOTCH and TP53 mutations. However, none of these genetic subtypes are associated with BCL2 gain or increased BCL2 mRNA expression. Thus, it can be inferred that the genetic features of the BCL2 super-expressor are distinct from genetic subtypes proposed by LymphGen classification or DLBCL cluster, which warrants further investigation in future studies.
This study has several limitations. As anticipated for any retrospective study, there may have been a selection bias. DHL data was not available for approximately half of the patients in the training set. However, this would not have significantly impacted the results as DHL was very rare in our cohort (2/136, 1.5%), consistent with the results of a recent study of Korean DLBCL patients46. Despite these limitations, the prognostic value of the BCL2 super-expressor status proposed in the present study was evaluated in a relatively large cohort and was confirmed in an independent validation set. As BCL2 super-expressor status is based on clear criteria with high reproducibility and has significant prognostic value, we believe it can be easily applied to routine clinical practice. In conclusion, this study suggests that the BCL2 super-expression characterizes a distinct subgroup of DLBCL with a poor prognosis.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Coiffier, B. et al. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N. Engl J Med 346, 235–242 (2002).
Vitolo, U. et al. Obinutuzumab or rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone in previously untreated diffuse large B-cell lymphoma. J Clin Oncol 35, 3529–3537 (2017).
Crump, M. et al. Outcomes in refractory diffuse large B-cell lymphoma: results from the international SCHOLAR-1 study. Blood 130, 1800–1808 (2017).
Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503–511 (2000).
Rosenwald, A. et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl J Med 346, 1937–1947 (2002).
Davies, A. et al. Gene-expression profiling of bortezomib added to standard chemoimmunotherapy for diffuse large B-cell lymphoma (REMoDL-B): an open-label, randomised, phase 3 trial. Lancet Oncol 20, 649–662 (2019).
Vitolo, U. et al. ROBUST: First report of phase III randomized study of lenalidomide/R-CHOP (R2-CHOP) vs placebo/R-CHOP in previously untreated ABC-type diffuse large B-cell lymphoma. Hematol Oncol 37, 36–37 (2019).
Younes, A. et al. Randomized phase III trial of ibrutinib and rituximab plus cyclophosphamide, doxorubicin, vincristine, and Prednisone in non-germinal center B-cell diffuse large B-cell lymphoma. J Clin Oncol 37, 1285–1295 (2019).
Chapuy, B. et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24, 679–690 (2018).
Schmitz, R. et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl J Med 378, 1396–1407 (2018).
Herrera, A. F. et al. Relapsed or refractory double-expressor and double-hit lymphomas have inferior progression-free survival after autologous stem-cell transplantation. J Clin Oncol 35, 24–31 (2017).
Johnson, N. A. et al. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 30, 3452–3459 (2012).
Green, T. M. et al. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 30, 3460–3467 (2012).
Hu, S. et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from the international DLBCL rituximab-CHOP consortium program. Blood 121, 4021–4031 (2013). quiz 4250.
Morschhauser, F. et al. A phase 2 study of venetoclax plus R-CHOP as first-line treatment for patients with diffuse large B-cell lymphoma. Blood 137, 600–609 (2021).
Riedell, P. A. & Smith, S. M. Double hit and double expressors in lymphoma: definition and treatment. Cancer 124, 4622–4632 (2018).
Czabotar, P. E., Lessene, G., Strasser, A. & Adams, J. M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 15, 49–63 (2014).
Hermine, O. et al. Prognostic significance of bcl-2 protein expression in aggressive non-Hodgkin’s lymphoma. Groupe d’Etude des Lymphomes de l’Adulte (GELA). Blood 87, 265–272 (1996).
Tsuyama, N. et al. BCL2 expression in DLBCL: reappraisal of immunohistochemistry with new criteria for therapeutic biomarker evaluation. Blood 130, 489–500 (2017).
Meyer, P. N. et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol 29, 200–207 (2011).
Roh, J. et al. Quantitative analysis of tumor-specific BCL2 expression in DLBCL: refinement of prognostic relevance of BCL2. Sci Rep. 10, 10680 (2020).
Ventura, R. A. et al. FISH analysis for the detection of lymphoma-associated chromosomal abnormalities in routine paraffin-embedded tissue. J Mol Diagn 8, 141–151 (2006).
Mossafa, H. et al. Non-Hodgkin’s lymphomas with Burkitt-like cells are associated with c-Myc amplification and poor prognosis. Leuk Lymphoma 47, 1885–1893 (2006).
Lohr, J. G. et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci USA 109, 3879–3884 (2012).
Miao, Y., Medeiros, L. J., Li, Y., Li, J. & Young, K. H. Genetic alterations and their clinical implications in DLBCL. Nat Rev Clin Oncol 16, 634–652 (2019).
Morin, R. D. et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476, 298–303 (2011).
Pasqualucci, L. et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet 43, 830–837 (2011).
Chen, C., Khaleel, S. S., Huang, H. & Wu, C. H. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol Med 9, 8 (2014).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31, 213–219 (2013).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010).
Benjamin, D. et al. Calling somatic SNVs and indels with Mutect2. bioRxiv https://doi.org/10.1101/861054 (2019).
Reddy, A. et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 171, 481–494 e415 (2017).
Lacy, S. E. et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood 135, 1759–1771 (2020).
Iqbal, J. et al. BCL2 predicts survival in germinal center B-cell-like diffuse large B-cell lymphoma treated with CHOP-like therapy and rituximab. Clin Cancer Res 17, 7785–7795 (2011).
Petrella, T. et al. BCL2 expression but not MYC and BCL2 coexpression predicts survival in elderly patients with diffuse large B-cell lymphoma independently of cell of origin in the phase 3 LNH03-6B trial. Ann Oncol 28, 1042–1049 (2017).
Kluk, M. J. et al. MYC immunohistochemistry to identify MYC-driven B-cell lymphomas in clinical practice. Am J Clin Pathol 145, 166–179 (2016).
Mahmoud, A. Z. et al. Scoring of MYC protein expression in diffuse large B-cell lymphomas: concordance rate among hematopathologists. Mod Pathol 28, 545–551 (2015).
Adams, C. M., Clark-Garvey, S., Porcu, P. & Eischen, C. M. Targeting the Bcl-2 family in B cell lymphoma. Front Oncol 8, 636 (2019).
Collinge, B. et al. The impact of MYC and BCL2 structural variants in tumors of DLBCL morphology and mechanisms of false-negative MYC IHC. Blood 137, 2196–2208 (2021).
Ennishi, D. et al. Genetic profiling of MYC and BCL2 in diffuse large B-cell lymphoma determines cell-of-origin-specific clinical impact. Blood 129, 2760–2770 (2017).
Huang, S. et al. Prognostic impact of diffuse large B-cell lymphoma with extra copies of MYC, BCL2 and/or BCL6: comparison with double/triple hit lymphoma and double expressor lymphoma. Diagn Pathol 14, 81 (2019).
Ott, G., Rosenwald, A. & Campo, E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood 122, 3884–3891 (2013).
Ye, Q. F. et al. Silencing Notch-1 induces apoptosis and increases the chemosensitivity of prostate cancer cells to docetaxel through Bcl-2 and Bax. Oncol Lett 3, 879–884 (2012).
Wright, G. W. et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 37, 551–568 (2020).
Han, B. et al. Immunophenotypic landscape and prognosis of diffuse large B-cell lymphoma with MYC/BCL2 double expression: an analysis of a prospectively immunoprofiled cohort. Cancers (Basel) 12, 3305 (2020).
Acknowledgements
This study was supported by a grant of the National Research Foundation of Korea (no. NRF-2020R1A2C2006362 to Chan-Sik Park; NRF-2021R1C1C2007816 to J.R., http://www.nrf.re.kr) and by grants from the Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea (no.18–527, no.2020IP0064, and no.2021IL0008 to Chan-Sik Park). However, these funding bodies had no role in the study design, data collection, analysis, publication decision, or manuscript preparation. The authors received no specific funding for this work.
Author information
Authors and Affiliations
Contributions
C.-S.P. and D.H.Y. conceived and designed the study; J.R. and H.C wrote the manuscript; C.-S.P., J.R., and J.H. made the histopathological diagnoses and analyzed the immunohistochemical data; J.R., H.C., Y.S.L., S.-W.K., J.-S.R., E.J.C., K.W.K., J.H., Y.S.C., S.H.J., C.S., D.H.Y., and C.-S.P. contributed to the patient care and collected the clinical data; J.R., H.C., H.-K.P., Y.S.C., C.S., D.H.Y. and C.-S.P. analyzed the results. All authors had full access to the data, carefully reviewed the manuscript, and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Roh, J., Cho, H., Pak, HK. et al. BCL2 super-expressor diffuse large B-cell lymphoma: a distinct subgroup associated with poor prognosis. Mod Pathol 35, 480–488 (2022). https://doi.org/10.1038/s41379-021-00962-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41379-021-00962-z
This article is cited by
-
The role of comorbidity indices and histochemical markers in surgically resected and non-resected primary central nervous system lymphoma
Clinical and Experimental Medicine (2023)